Sasaki et al. 10.1073/pnas.0603182103. |

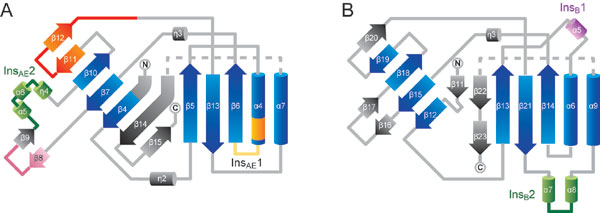
Fig. 6. The B3/4 domain architectures. (A and B) Diagrams of the B3/4 domain secondary structures from P. horikoshii (A) and T. thermophilus (B) PheRS, colored as in Fig. 1 B and C.

Fig. 7. A superposition of the B1 and B5 domains. (A and B) The rotation of the B3/4 domain. Ribbon models of P. horikoshii PheRS-bN (A) and the T. thermophilus counterpart (B) are shown in the direction that superimposes both the B1 and B5 domains, colored as in Fig. 1A. InsB1 is colored purple. (C and D) Model for the CCA end of Tyr-tRNAPhe translocation. Based on T. thermophilus PheRS complexed with tRNAPhe (C), tRNAPhe was placed in the P. horikoshii PheRS-bN (D). The CCA end (shown in orange) was modeled to the editing pocket. Yellow, tRNAPhe; Gray, PheRS a-subunit. The B2 domain in T. thermophilus PheRS is not shown.

Fig. 8. Posttransfer editing by the P. horikoshii PheRS mutants that are not shown in Fig. 4A. Tyr-tRNAPhe deacylation by the WT and mutant PheRSs is shown. The graph is the same experiment as in Fig. 4A. Reactions were performed with 10 nM PheRSs.
Table 1. Crystallographic data and refinement statistics
| SeMet | ||
Peak | Edge | Remote | |
Data collection |
|
|
|
Space group | C 2 |
|
|
Unit cell dimensions | a = 142.8 Å |
|
|
b = 82.0 Å |
|
| |
c = 139.1 Å |
|
| |
β = 116.8° |
|
| |
Wavelength, Å | 0.9793 | 0.9796 | 0.9840 |
Resolution, Å | 50.00-1.94 (2.01-1.94)* | 50.00-2.00 (2.07-2.00)* | 50.00-2.00 (2.07-2.00)* |
Unique reflections | 104,810 | 96,882 | 96,964 |
Completeness, % | 99.4 (94.2)* | 100.0 (100.0)* | 100.0 (100.0)* |
R sym (%)† | 12.3 (63.9)* | 12.1 (56.8)* | 11.7 (57.1)* |
Redundancy | 7.0 (4.9)* | 3.7 (3.7)* | 3.8 (3.7)* |
I /σ(I) | 18.6 (1.91)* | 11.8 (1.68)* | 13.1 (1.89)* |
|
|
| |
Refinement statistics |
|
|
|
Resolution, Å | 50.00-1.94 |
|
|
No. reflections | 99,536 |
|
|
R work/Rfree‡ | 0.188/0.231 |
|
|
No. protein atoms | 8,544 |
|
|
No. water molecules | 780 |
|
|
Average B factor, Å2 | 26.3 |
|
|
Rmsd values |
|
|
|
Bond length, Å | 0.017 |
|
|
Bond angles, ° | 1.546 |
|
|
Ramachandran plot, % |
|
|
|
Favored | 92.0 |
|
|
Additional allowed | 7.5 |
|
|
Generously allowed | 0.5 |
|
|
Disallowed | 0.0 |
|
|
*The statistics in the highest-resolution shell are given in parentheses.
†
Rsym = Σ|I - <I>|/ΣI, where I is the observed intensity of reflections.‡
Rwork, free = Σ|Fobs - Fcalc|/ΣFobs. Free reflections consist of 5% of the total number of reflections.Supporting Text
Sequence Analysis
A total of 193 phenylalanyl-tRNA synthetase (PheRS)-b sequences (8 from eukarya, 18 from archaea, and 167 from bacteria) were retrieved from the SWISS-PROT database. Because the bacterial sequences from Treponema pallidum and Borrelia burgdorferi have no bacteria-specific features, these were removed from the sequence set. We also removed redundant sequences with >90% amino acid similarity, according to Gonnet PAM 250. The archaeal/eukaryal set (25 sequences, 7 from eukarya and 18 from archaea) and the bacterial set (116 sequences) were independently aligned with the ClustalX program, version 1.83 (1). Then, the two alignments were merged and corrected manually, based on the 3D structures of the Pyrococcus horikoshii and Thermus thermophilus PheRS-bs.
The Docking Model
To build the model of the P. horikoshii editing domain with 2'-tyrosylated adenosine (Tyr-A76), we used the structure of the T. thermophilus B3/4 domain with the bound tyrosine molecule as a reference (Protein Data Bank entry 2AMC). Based on the structure-based alignment (Fig. 2), the P. horikoshii B3/4 domain (residues 102-116, 121-132, 141-169, 198-205, 209-217, 222-236, 242-256, and 258-261) was superimposed on the T. thermophilus counterpart (residues 211-225, 244-255, 259-287, 296-303, 310-318, 322-336, 368-382, and 385-388, respectively). From this superposition, a draft model of the P. horikoshii B3/4 domain with tyrosine was made. Then, we attached the tRNA 3'-terminal adenosine moiety (A76) to the tyrosine. Finally, the total energy of the model was minimized using cns (model_minimize.inp; ref. 2) for 200 steps with all of the atoms nonfixed.
1. Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acids Res 22:4673-4680.
2. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Acta Crystallogr D 54:905-921.