Supplementary material for Hassanin et al. (2000) Proc. Natl. Acad. Sci. USA 97 (21), 11415-11420.

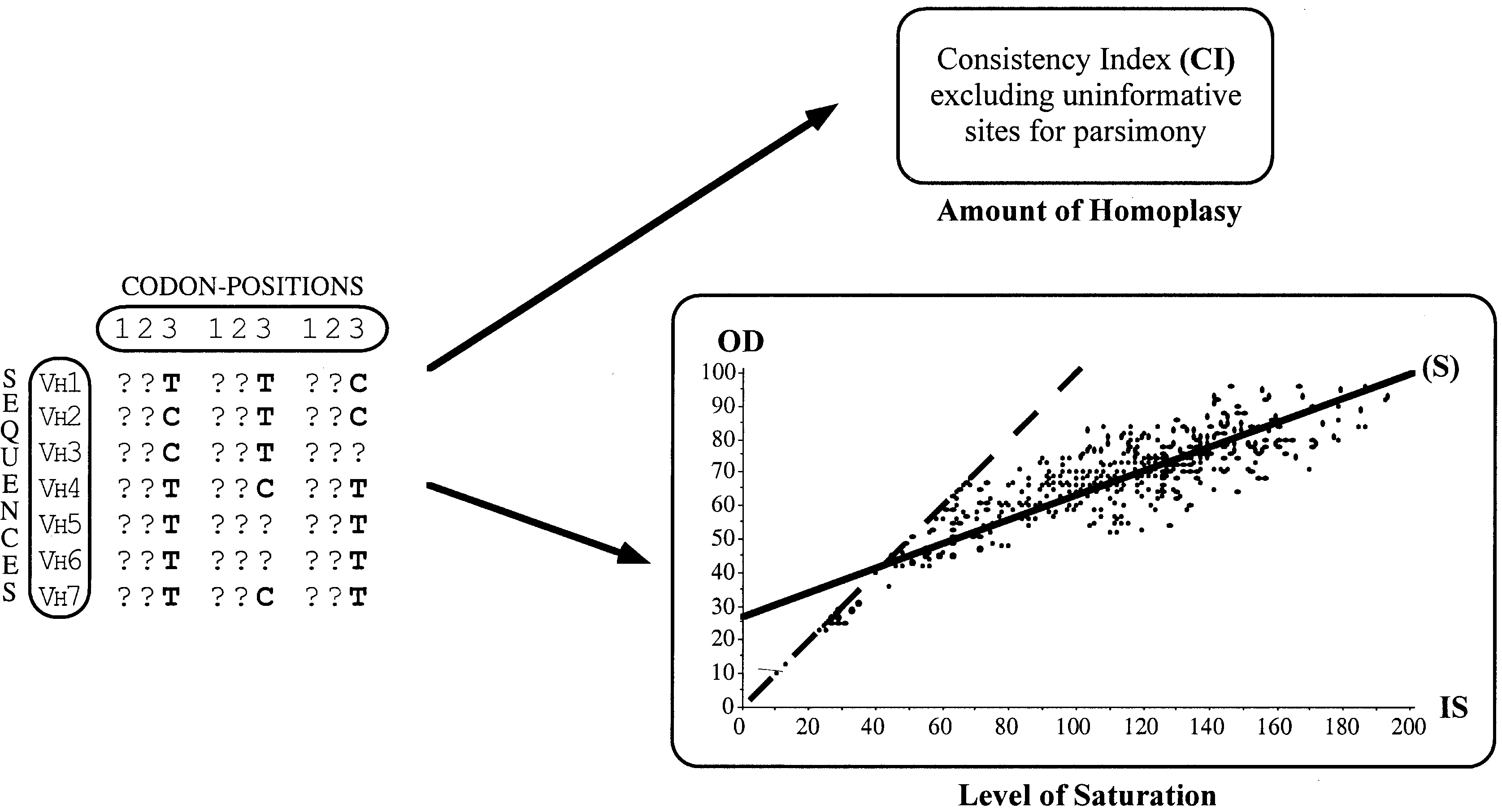
Fig. 6. Two approaches for measuring homoplasy of C-T substitutions at third codon positions. To analyze the level of homoplasy for C-T substitutions at third codon positions, nucleotides A and G have been replaced by question marks in the original DNA alignment, and only third codon positions have been selected for parsimony analysis. Two approaches were used to estimate the homoplasy content. (i) The value of consistency index (CI) excluding uninformative sites was given by PAUP ("Describe Trees" option). (ii) The analysis of saturation was assessed by plotting the pairwise number of observed differences (OD) against the corresponding pairwise number of inferred substitutions (IS) for one of the most-parsimonious trees obtained with PAUP ("Patristic Distances" option). The slope of the linear regression (S) was used as an index representing the level of saturation. The dotted line represents the area of equal numbers of observed and inferred changes, i.e., the theoretical situation for which no saturation is observed.