
Westlake et al. 10.1073/pnas.0610500104. |

Fig. 5. Evi5 interacts with the constitutively active form of Rab11a-Q70L by yeast two-hybrid analysis. Bait (Evi5N R208A) and prey (Rab-QL) constructs were cotransformed into AH109 yeast and plated first on double selection (-Leu, Trp) followed by quadruple selection (-Leu, Trp, Ade, His) plates.

Fig. 6. Alignment of the TBC domains from Evi5, candidate Evi5 orthologs, and GAPCenA. The absolutely conserved IxxDxxR motif required for GAP catalytic activity is indicated. Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis; Dr, Danio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana.

Fig. 7. Evi5 does not display GAP activity toward Rab11 in vitro. (A) Wild-type Rab11a displays significant intrinsic GTPase activity, whereas Rab11a-Q70L is GTPase-deficient. TLC analysis was performed as described for the GAP assay with the exception that only GST-Rab proteins were included in the reaction. Data were normalized to account for GDP present before MgCl2 addition at time 0. (B) Evi5 does not stimulate Rab11 GTPase activity as measured by the TLC method. Bacterially expressed GST-Rab proteins were preloaded with [a-32P]GTP and incubated with MBP or MBP-Evi5N protein at 30°C. This panel shows the raw data that are quantified and shown in Fig. 1D. (C) Evi5 does not stimulate Rab11 GTPase activity as measured by detection of inorganic phosphate release. MBP-Evi5N and Rab11b, or either protein on its own, was incubated for 1 h and inorganic phosphate generation was measured. Stimulation of GTP hydrolysis by p50-RhoGAP was used as a positive control.

Fig. 8. Rab11 overexpression localizes Evi5 to Rab11-positive endosomes. (A) Localization of GFP-tagged Rabs and Myc-Evi5 in singly transfected cells. U2OS cells were transfected with GFP-Rab or Myc-Evi5 for 48 h, then fixed with 4% PFA and processed for immunofluorescence analysis with anti-Myc antibody (Myc-Evi5 treatment only) and Hoechst to mark the DNA. (B) Expressed wild-type and constitutively active Rab11 recruits Evi5 to Rab11-positive endosomes. U2OS cells were transfected with Myc-Evi5 and various GFP-Rab constructs, fixed with 4% PFA, and stained with anti-Myc antibody as well as Hoechst to mark the DNA. White arrows highlight sample regions of colocalization, which appear yellow in the merged panel.
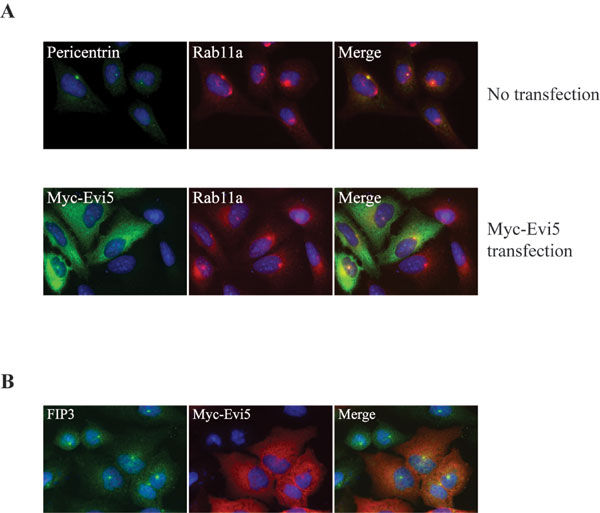
Fig. 9. Overexpression of Evi5 does not alter localization of Rab11 or the Rab11 effector FIP3. (A) Rab11 localizes normally after Evi5 overexpression. (Upper) Untransfected U2OS cells were fixed with 4% paraformaldehyde and stained with Hoechst as well as antibodies against pericentrin (to mark the centrosome) and Rab11. (Lower) U2OS cells were transiently transfected with a Myc-Evi5 full-length construct for 48 h, fixed in 4% paraformaldehyde, and stained with anti-Myc and anti-Rab11a antibodies. (B) FIP3 localizes normally in the presence of Evi5 overexpression. HeLa cells were transiently transfected with Myc-Evi5 full-length for 48 h, fixed in 4% paraformaldehyde, and stained with anti-Myc antibody and anti-FIP3 antibody, as well as Hoechst to mark the DNA.

Fig. 10. RabGAPs and Rab effectors are predicted to occupy a similar space when bound to Rab protein. Solved structures of a single Rab11a-Q70L molecule (green) in complex with a FIP2 dimer (red) in the presence of GTP and Mg (Left) and the yeast RabGAP Gyp1 (cyan) bound to human Rab33 (yellow) in the presence of GDP and AlF3 (Center) are shown. Structural representations were modified from Protein Data Bank entries 2GZD (1) and 2G77 (2), and alignment and overlay of the Rab complexes was performed by using PyMOL (Right). GTP (purple) and GDP (magenta) are shown.
1. Pan X, Eathiraj S, Munson M, Lambright DG (2006) Nature 442:303-306.
2. Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ, McCaffrey MW, Khan AR (2006) Structure (London) 14:1273-1283.

Fig. 11. RIP11/FIP5 competes with Evi5 for binding to Rab11a. Glutathione-Sepharose beads were incubated with GST-Rab11a GMP-PNP and then washed. The Rab11a beads were then incubated with a fixed amount of MBP-Evi5N protein (5 mg) and an increasing molar ratio of a carboxyl-terminal fragment of RIP11/FIP5, consisting of the Rab binding domain (either wild type or I629E point mutant). After washing the beads, bound proteins were eluted with SDS sample buffer and analyzed by SDS/PAGE followed by Coomassie staining.
SI Materials and Methods
Yeast Two-Hybrid Studies.
Rabs used in yeast two-hybrid studies were amplified by PCR using either full-length Rab cDNA clones (Invitrogen) or cDNA libraries as template and cloned into pGBKT7 (Clontech, Mountain View, CA). Each Rab was engineered to remove the carboxyl-terminal prenylation site, and site-specific mutations (active site Gln to Leu) were introduced to generate constitutively active, GTP-bound Rabs using QuikChange (Stratagene, La Jolla, CA). Evi5N (amino acids 1-376) R208A was subcloned into pGADT7 (Clontech) using standard methods. Positive interactions (AH109 yeast strain) were identified by selection on plates lacking leucine, tryptophan, adenine, and histidine.Bacterial Expression and Purification of Rabs.
GST-Rab expression constructs were transformed into BL21 DE3 RIII or BL21 DE3 pLysS E. coli strains and protein production was induced with IPTG overnight at 25°C. Pelleted bacteria were lysed in either PBS (pH 7.4)/0.1% Triton X-100 or 20 mM Hepes (pH 7.4)/500 mM NaCl/0.5% Triton X-100 buffers containing 1 mM DTT and Complete protease inhibitors (Roche, Indianapolis, IN). Lysates were cleared by centrifugation at 30,000 ´g for 30 min at 4°C. When preparing specifically active or inactive Rab proteins, GMP-PNP or GDP nucleotides were included in buffers throughout the purification protocol. Supernatants containing GST fusion proteins were incubated with glutathione-Sepharose beads (Sigma, St. Louis, MO) for 1 h at 4°C. Beads were washed extensively in lysis buffer and eluted in buffer (either PBS, pH 7.4/2.5 mM MgCl2 or 20 mM Tris, pH 7.5/2.5 mM MgCl2/1 mM DTT) plus 10 mM reduced glutathione. For binding assays eluates containing protein were pooled and dialyzed against PBS (pH 7.4)/2.5 mM MgCl2.Sequence and Structural Alignment.
Primary sequence alignments were performed by using ClustalW. Protein structures were visualized and aligned with PyMOL.GAP Assay Using Inorganic Phosphate Detection.
The RhoGAP inorganic phosphate detection assay was carried out according to the manufacturer's instructions (Cytoskeleton, Denver, CO). Briefly, GST-Rab and MBP-Evi5N protein (both at 5 mM) were incubated for 1 h at 37°C with 200 mM GTP. Absorbance readings were measured at 650 mm.