Rizvi and Raghaven 1010.1073/pnas.0605131103 |
Fig. 6. Expression of sTpn in CHO cells and protein purification. (A) Chinese hamster ovary cells (CHO) cells were transfected with a construct encoding soluble Tpn (sTpn) in the pBJ5-GS vector and selected with 100 mM L-methyl sulfoximine (MSX). Cells surviving selection were metabolically labeled with [35S]methionine/cysteine for 5 h, followed by immunoprecipitations (IP) of cell supernatants with an anti-FLAG antibody. Lane 1, pBJ5-GS-sTpn transfected and MSX-selected cells; lane 2, untransfected cells. Proteins were visualized by SDS/PAGE and phosphorimaging analyses. A 46- to 48-kDa protein was detectable in these analyses, and cells expressing this component were clonally expanded. (B-D) CHO cell lines expressing sTpn were further subcloned and screened for clones expressing sTpn by the assay described in A. For protein purification, cells were grown to confluence in 10-cm2 plates and media was collected every second or third day. sTpn was purified from supernatants using anti-FLAG affinity beads (Sigma). sTpn was eluted from the column using 100 mg/ml FLAG peptide (N-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-C) in 50 mM Tris and 150 mM NaCl, pH 7.5. The protein was concentrated and further purified by gel filtration chromatography (Superose 6, Amersham Pharmacia) in 50 mM Tris and 150 mM NaCl, pH 7.5 (B and C). Monomers were predominant in sTpn purified from supernatants of CHO cells cultured at 30-33°C (B), whereas parallel culturing of cells at 37°C led to increased aggregation (C). In preparations from cells cultured at 37°C (C), peak fractions corresponding to monomeric forms of sTpn were collected, pooled, and repurified by gel filtration chromatography before use in binding experiments. These second gel filtration chromatograms always showed a single peak corresponding to the monomeric forms of Tpn (D). E) Anti-FLAG and gel filtration chromatography-purified sTpn resolved by SDS/PAGE and Coomassie blue staining.
Fig. 7. Peptide loading of A2 or of B35 purified from insect cell supernatants. Native-PAGE and fluorimaging analyses of peptide binding by A2 and B35 in the presence or absence of sTpn. Soluble A2 and B35 heterodimers were purified from insect cells for these analyses. Since insect cells lack class I assembly factors, expression in insect cells allows for purification of peptide-deficient class I (1, 2). Indicated proteins were incubated at 37°C (in 50 mM Tris, 150 mM NaCl, pH 7.4) for the indicated amounts of time. Insets show representative gel panels following fluorimaging analyses, and graphs show the quantifications of relative peptide binding. Binding of high affinity (LLDCFPTAAV) and low affinity (LLDCFPTAAVQ) A2 peptides, and binding of high affinity (LPSCFADVEF) B35 peptides were undertaken. (A) Simultaneous incubation of A2 ± sTpn (2.5 mM each) and LLDCFPTAAV (0.08, 2.5 or 25 m M) at 37°C for 30/60 min, or A2 alone for 240 or 360 min to demonstrate that binding was not saturating during the 30/60 min incubations. Graphs are average of three independent analyses. (B) B35 ± sTpn (2.5 mM each) were simultaneously incubated at 37°C with LPSCFADVEF (0.08, 2.5 mM) for 60 or 240 min. The longer (240 min) time point of B35 + LPSCFADVEF incubation showed that binding at the 60 min time point was not saturating (with 0.08 mM peptide). The lower panel shows binding data averaged over three independent analyses. (C and D) Simultaneous incubation of A2 ± sTpn with LLDCFPTAAVQ at 2.5 mM (C) or at 25 mM (D) peptides for the indicated times. A2 + LLDCFPTAAV was included to demonstrate reduced relative binding of LLDCFPTAAVQ, that has a C-terminal residue extension predicted to reduce A2 binding affinity. Graphs are average of two independent analyses.
Fig. 8. At low class I concentrations, b2m markedly enhances peptide loading of class I. Native-PAGE and fluorimaging analyses of peptide binding by A2 and B35 heterodimers in the presence of b2m (1 mM) or buffer. Indicated proteins were incubated at 37°C (in 50 mM Tris, 150 mM NaCl, pH 7.4) with 1 m M LLDCFPTAAV (for A2) or LPSCFADVEF (for B35). (Upper) Representative gel panels following fluorimaging analyses. (Lower) Graphs show the quantifications of relative peptide binding. Graphs are averages of three independent analyses.
Fig. 9. Peptide loading of empty class I. Native-PAGE and fluorimaging analyses of peptide binding by empty A2 (eA2) and empty B35 (eB35), in the presence or absence of sTpn and control protein BSA (BSA). (A and B) eA2 or eB35 (0.1 mM) were incubated with 0.05 mM LLDCFPTAAV (for A2) or 0.05 mM LPSCFADVEF (for B35) and b2m (0.5-1 mM) in the presence of sTpn (0.5 mM), BSA (0.5 mM), or buffer at room temperature for indicated times. The peptide binding signals were quantified following Native-PAGE and fluorimaging analyses. Data are average of two independent analyses (A) or four independent analyses (B). Only a small increase in peptide loading was observed in the presence of sTpn compared to the BSA control. (C and D) eA2 or eB35 (0.1 mM each) were incubated with indicated amounts of peptide, b2m (1 mM) and BSA (8-10 mM) in the presence of sTpn (0.5 mM) or additional BSA (0.5 mM) at room temperature for 10 min. The peptide binding signals were quantified following Native-PAGE and fluorimaging analysis and are shown as bar graphs. Data are averaged over two analyses and representative of 4 and 2 (C and D) independent analyses, respectively. An increase in peptide binding was observed in samples containing sTpn and class I relative to BSA and class I, but the effects were small, and not always reproducible in other experiments. (E and F) To examine peptide loading of class I under assembly-limiting conditions, eA2 or eB35 (0.1 mM each), were incubated with indicated peptide (without excess b2m) and excess BSA (8-10 mM) in the presence of sTpn (0.5 mM) or additional BSA (0.5 mM) at room temperature for 10 min. The peptide binding signals were quantified following Native-PAGE and fluorimaging analysis and are shown as bar graphs. Data are averaged over two analyses and are representative of three or four independent analyses, respectively.
Fig. 10. sTpn-empty class I complexes are assembly competent. Empty class I alone, or sTpn and empty class I were incubated at 30°C for 6 h, loaded over anti-FLAG beads, and eluted with FLAG peptide (A and C Upper, lanes 1 and 2). The class I alone incubations served as negative controls, to assess non-specific binding to anti-FLAG beads. The amount of sTpn-associated class I was estimated by titration against known amounts of empty-class I (A and C Upper). The amount of A2 HC in complex with sTpn was estimated to be between 10-20 ng in the indicated sample volume (A Upper, lane 2 compared with 3 and 4), and the amount of B35 HC in complex with sTpn was estimated to be between 20-40 ng in the indicated sample volume (C Upper, lane 2 compared with 4 and 5). sTpn-associated A2 and B35 were both assembly competent, although the rates were reduced relative to the corresponding "free" empty proteins (compare Lower panels of A, lanes 2 and 4, and C lanes 2 and 5, and corresponding quantifications in B and D). The sTpn-complexed proteins were subject to several treatments compared to free class I, including a 30°C incubation for 6 h (to maximize complex formation), and purification using anti-FLAG beads, which may have depleted any b2m that was not associated with sTpn/class I. Because of these differences in experimental conditions for maintenance of sTpn-associated and free proteins, precise quantitative comparison of relative peptide binding by the free and sTpn-associated samples is not possible. Nevertheless, these analyses illustrated that a significant fraction of the class I protein purified as a complex with sTpn on anti-FLAG beads was competent for subsequent assembly with peptide.

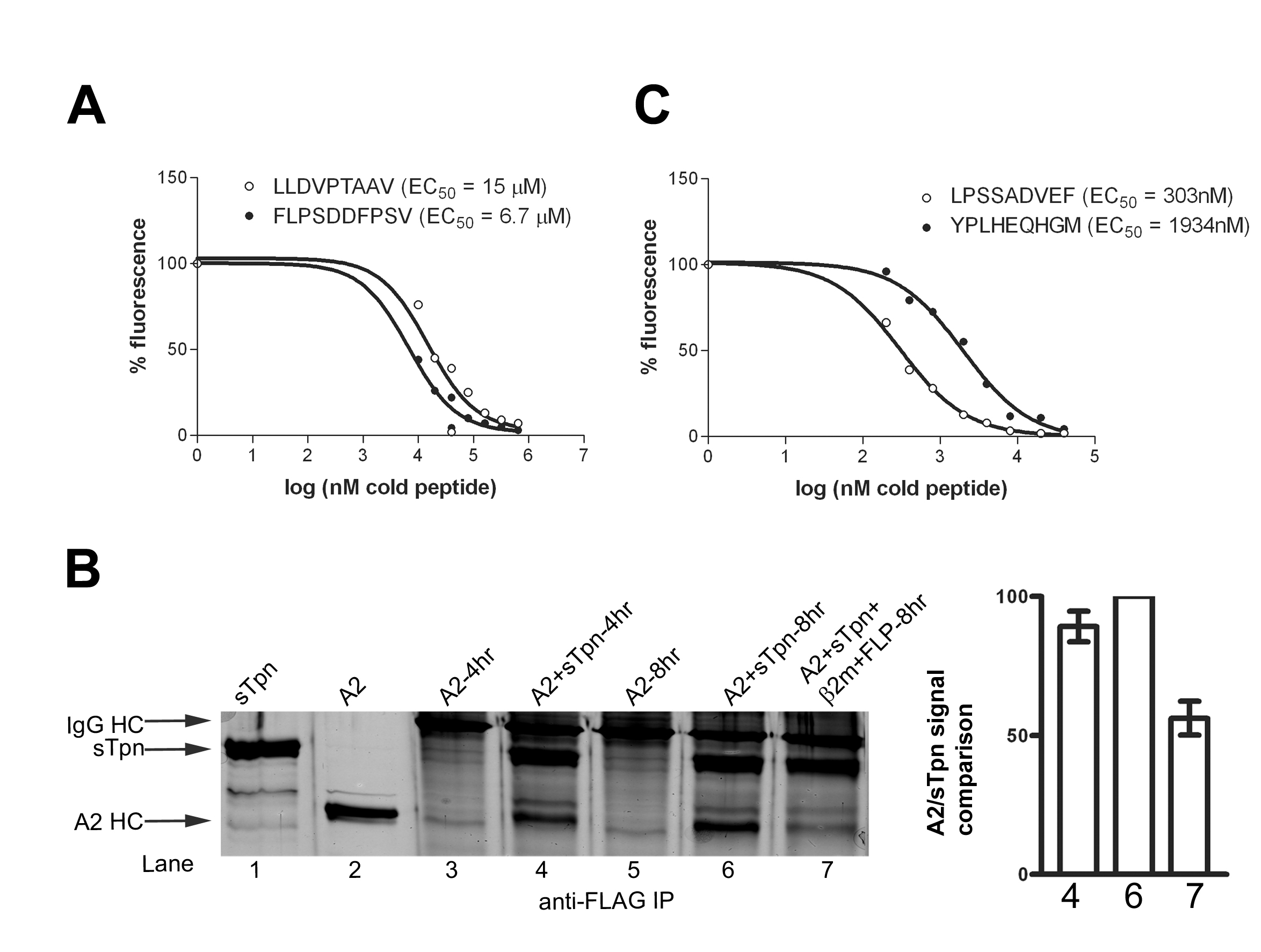
Fig. 11. Relative affinities of A2 and B35 specific peptides and FLPSDDFPSV-induced dissociation of A2-sTpn. (A and C) A2 or B35 (2 mM) were incubated with fluorescently labeled peptide LLDCFPTAAV or YPLKFEQHGM (2 or 0.2 mM) in the presence of various concentrations of unlabeled peptides (LLDVPTAAV or FLPSDDFPSV for A2 and LPSSADVEF or YPLHEQHGM for B35) at 37°C for 1 h. The peptide binding signals were quantified following Native-PAGE and fluorimaging analysis, and inhibition data were analyzed using Prism to calculate the indicated EC50 values. (B Left) Silver-stained SDS-PAGE gels of indicated proteins (lanes 1 and 2) or anti-FLAG IP (lanes 3-7). IPs were of A2 and sTpn (5 mM each) incubated for 4 h at 37°C (lane 4), followed by additional 4 h incubation with buffer (lane 6), or with 50 mM b2m and 500 mM FLPSDDFPSV (FLP) (Lane 7). (Right) Bar graphs shows class I/sTpn intensity ratio comparisons for the indicated lanes, with the highest ratio set at 100%. Data in Right panels are A2/sTpn intensity ratios from the indicated lanes averaged over five independent analyses.
Fig. 12. High affinity peptides are better inhibitors of the sTpn-class I interaction than low affinity peptides. (A-C Left) Silver-stained SDS/PAGE gels. Anti-FLAG IP were undertaken following protein or buffer incubations at 37°C. (A-C Right) Bar graphs show class I/sTpn intensity ratio comparisons for the indicated lanes, with the highest ratio in each gel set at 100%. (A) Anti-FLAG IP of A2 (lane 1, 2.5 mM), A2 + sTpn (2.5 mM each) ± peptide as indicated (lanes 2-6) or sTpn alone (lane 7, 2.5 mM). A background band that appears in all lanes is indicated by an asterisk. (B and C) The lanes contained direct protein loads of sTpn (lane 1, 0.25 mg), B35 (lane 2, 0.25 mg) or various IP (lanes 3-8). B35 was simultaneously incubated at 37°C for 2 h ± sTpn, and ± LPSCFADVEF (B) or YPLHEQHGM (C) as indicated, before the IP. Data in Right panels are averaged over two (A and C) or three (B) independent analyses.
Supporting Materials and Methods
Protein Expression and Purification.
Class I heterodimers from insect cell supernatants were purified using W6/32 affinity chromatography (1, 2), and further purified by gel filtration chromatography. Human b2m (3) or chimp b2m (identical in sequence to human b2m) ligated into the NdeI and BamHI sites of the pET3a vector (Novagen) were expressed in E. coli, purified as inclusion bodies, and solubilized and refolded using established procedures. sTpn was amplified using the following primers:5'-ATGGATCCGCGGCCGCCCACCATGAAGTCCCTGTCTCTGCTCCTC-3' and 5'-ATGGATCCGCGGCCGCTTATTTATCATCATCATCTTTATAATCGTTCTCA AGGGAGGGCCCTGAAA-3' using full-length Tpn in PCR-Script as template (4). The reverse primer encodes a FLAG peptide sequence before the stop codon. NotI sites were inserted flanking the Tpn sequence, and a stop codon was inserted following that corresponding to amino acid 392 of Tpn, which truncates the Tpn protein before the start of its transmembrane sequence. The PCR product was digested with NotI and ligated into NotI site of pBJ5-GS vector (5). pBJ5-GS-Tpn was transfected into CHO cells using a Lipofectin procedure (Life Technologies, Inc.) and CHO cells resistant to 100 mM L-methyl sulfoximine were selected as described previously (5). sTpn was purified from CHO cells using anti-FLAG affinity-column (Fig. 6).
Coimmunoprecipitation Analyses in Cells.
Insect cells (2 ´ 107) (Sf21) were infected with appropriate baculoviruses at 26°C at desired multiplicity of infection. Forty-eight hours postinfection, the cells were metabolically labeled with [35S]methionine/cysteine for 1 h, lysed and immunoprecipitated as described previously (4), but using anti-His antibody to detect A2 and M10, or PaSta-1 antibody (obtained from P. Cresswell, Yale University School of Medicine, New Haven, CT) to detect Tpn. Immunoprecipitated proteins were washed (10 mM phosphate buffer/10 mM Tris/130 mM NaCl/1% Triton-X100, pH 7.5), resolved by SDS/PAGE, and gels were exposed to a PhosphorImager plate. Images were processed using Molecular Dynamics PhosphorImager SI software.1. Mancino L, Rizvi SM, Lapinski PE, Raghavan M (2002) Proc Natl Acad Sci USA 99:931-936.
2. Rizvi SM, Mancino L, Thammavongsa V, Cantley RL, Raghavan M (2004) Mol Cell 15:913-923.
3. Garboczi DN, Hung DT, Wiley DC (1992) Proc Natl Acad Sci USA 89:3429-3433.
4. Raghuraman G, Lapinski PE, Raghavan M (2002) J Biol Chem 277:41786-41794.
5. Gastinel LN, Simister NE, Bjorkman PJ (1992) Proc Natl Acad Sci USA 89:638-642.