Palomero et al. 10.1073/pnas.0606108103. |

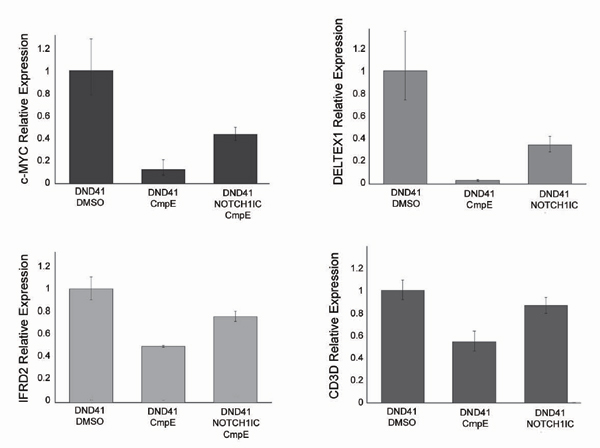
Fig. 7. ICN1 expression antagonizes the effects of GSIs on the expression of NOTCH1 target genes. Control DND41 T-ALL cells expressing GFP and DND41 cells infected with retroviruses driving expression of ICN1 and GFP were treated with GSI (CompE 100 nM 48 h). Expression of NOTCH1 target genes c-MYC, DELTEX1, IFRD2 and CD3D was up-regulated in ICN1 expressing cells compared to controls. GAPDH mRNA levels were used as normalization control. Data shows mean and SD of triplicate measurements.

Fig. 8. Inhibition of NOTCH1 signaling with GSIs impairs cell cycle progression in T-ALL cells. CUTLL1 and DND41 cells were treated with GSI (CompE 100 nM) or vehicle only (DMSO) for 72 h. Cell cycle was analyzed by flow cytometry analysis of DNA content after PI staining. A reduction in proliferation with accumulation of cells in G1 was observed. Histograms show representative results of triplicate experiments. Similar results were obtained in HPB-ALL, ALL-SIL, KOPTK1, and TALL1 cells.

Fig. 9. Venn diagrams representing the overlap between NOTCH1 direct target genes identified by ChIP-on-chip and genes regulated by GSI treatment in T-ALL cell lines. a. Overlap with ChIP-on-chip targets with error model P-value cutoff <0.05. (b) Overlap with ChIP-on-chip targets with error model P-value cutoff <0.001. The significance of the overlap was assessed using Fisher's exact test.
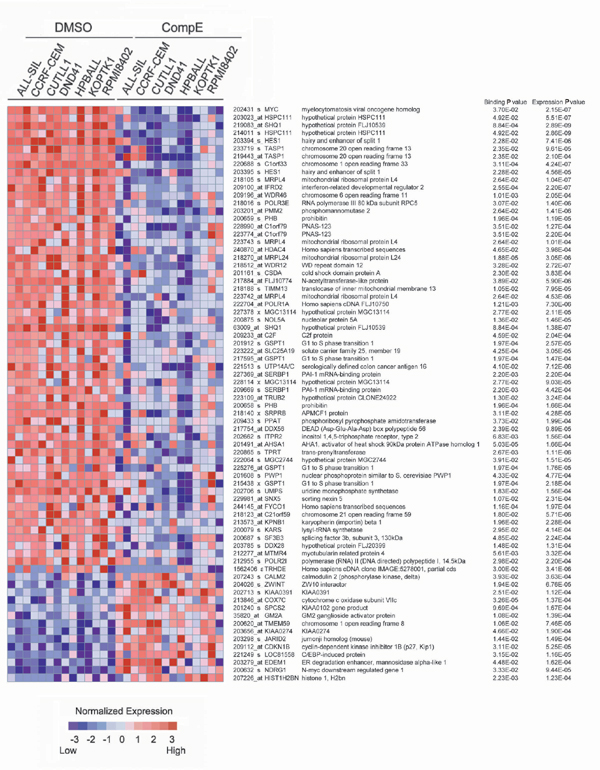
Fig. 10. Gene expression changes induced by GSI in genes identified as NOTCH1 targets by ChIP-on-chip. Genes with error model binding P values <0.05 and expression t test P values <0.001 are shown ranked by signal-to-noise ratio. Heat map represents color coded expression levels for each sample with respect to mock treatment controls.

Fig. 11. Structure of the proposed feed-forward loop regulatory motif controlling cell growth genes downstream of NOTCH1 and MYC in T-ALL. Input in the circuit is provided by activation of NOTCH1 signaling (by interaction of the NOTCH1 receptor with DSL ligands or by ligand independent activation induced by NOTCH1 mutations) and by signaling pathways up-regulating the expression or the activity of the MYC oncoprotein. This model support s that the intensity of input signals that enter this circuitry upstream of MYC may tune the cell growth response to NOTCH1 and may potentially enhance or attenuate the antitumor effect of drugs targeting NOTCH1 signaling.
Table 1. Top NOTCH1 direct target genes identified by ChIP-on-chip
Gene | Spot name | P value | Binding ratio | RefSeq no. |
PRCC | 4.76E-09 | 4.148045 | NM_005973 | |
BUB3 | BUB3 | 3.12E-08 | 3.44207 | NM_004725 |
FAM96A | FLJ22875 | 6.57E-08 | 3.331196 | NM_032231 |
PL6 | PL6 | 7.37E-08 | 3.33461 | NM_007024 |
PAFAH2 | PAFAH2 | 9.78E-08 | 3.393245 | NM_000437 |
RANGAP1 | RANGAP1 | 2.23E-07 | 3.227676 | NM_002883 |
TARBP2 | TARBP2 | 3.01E-07 | 3.093956 | NM_004178 |
ZNF133 | ZNF133_0,0 | 3.22E-07 | 3.041622 |
|
ZNF10 | ZNF10_0,0 | 3.34E-07 | 3.153393 |
|
HPS6 | FLJ22501 | 3.5E-07 | 2.930667 | NM_024747 |
TMEM48 | FLJ10407 | 5.76E-07 | 2.944545 | NM_018087 |
ING3 | ING3 | 6.53E-07 | 2.8508 | NM_019071 |
MDH1 | MDH1 | 9.83E-07 | 2.839357 | NM_005917 |
ZNF361 | ZNF361 | 1.17E-06 | 2.861604 | NM_018555 |
RC74 | FLJ10871 | 2.41E-06 | 2.710062 | NM_018250 |
FLJ10342 | 2.45E-06 | 2.726724 | NM_018064 | |
CFL1 | CFL1 | 2.51E-06 | 2.596959 | NM_005507 |
LTA4H | LTA4H | 2.6E-06 | 2.778543 | NM_000895 |
AAMP | AAMP | 2.96E-06 | 2.645006 | NM_001087 |
CPE | CPE | 3.37E-06 | 2.645677 | NM_001873 |
ZNF331 | BC009433_0,0 | 3.64E-06 | 2.663466 |
|
ACTR1A | ACTR1A | 3.65E-06 | 2.625543 | NM_005736 |
Cep 290 | FLJ13615 | 3.87E-06 | 2.657315 | NM_025114 |
SNX1 | SNX1 | 4.14E-06 | 2.608905 | NM_003099 |
MDS025 | MDS025 | 4.39E-06 | 2.798992 | NM_021825 |
TARBP2 | TARBP2_0,1 | 4.69E-06 | 2.52562 |
|
PPP1R12B | PPP1R12B | 5.26E-06 | 2.550444 | NM_032105 |
DDX23 | U5-100K | 6.8E-06 | 2.552142 | NM_004818 |
ZNF337 | ZNF337_0,0 | 8.26E-06 | 2.436899 | ? |
NPR2L | NPR2L | 8.31E-06 | 2.564293 | NM_006545 |
SKD3 | SKD3 | 1.23E-05 | 2.447516 | NM_030813 |
TRAP150 | TRAP150 | 1.25E-05 | 2.529101 | NM_005119 |
IFRD1 | IFRD1 | 1.39E-05 | 2.431396 | NM_001550 |
ZNF547 | NM_173631_0,0 | 1.47E-05 | 2.341599 |
|
SCNM1 | MGC3180 | 1.77E-05 | 2.52025 | NM_024041 |
HSPC138 | HSPC138 | 1.8E-05 | 2.431735 | NM_016401 |
MRPL24 | MRPL24 | 1.88E-05 | 2.527329 | NM_024540 |
USP5 | USP5 | 1.92E-05 | 2.402084 | NM_003481 |
DNAJC17 | FLJ10634 | 1.95E-05 | 2.437994 | NM_018163 |
ACAD8 | ACAD8 | 2.12E-05 | 2.339718 | NM_014384 |
PVRL2 | PVRL2 | 2.12E-05 | 2.571582 | NM_002856 |
UBE2V1 | UBE2V1 | 2.28E-05 | 2.902147 | NM_022442 |
ZNF225 | ZNF225 | 2.29E-05 | 2.513603 | NM_013362 |
SNAPC3 | SNAPC3_0,0 | 2.3E-05 | 2.341246 |
|
INVS | INVS | 2.31E-05 | 2.461536 | NM_014425 |
PDE6D | PDE6D | 2.43E-05 | 2.432897 | NM_002601 |
GTF2H4 | GTF2H4_0,0 | 2.59E-05 | 2.345813 |
|
LOC55815 | 2.86E-05 | 2.373108 | NM_018430 | |
COX7C | COX7C | 3.26E-05 | 2.254306 | NM_001867 |
FLJ12707 | 3.54E-05 | 2.309728 | NM_022067 | |
FTL | FTL | 3.66E-05 | 2.306027 | NM_000146 |
MGC4161 | MGC4161 | 4.03E-05 | 2.279359 | NM_024303 |
PEMT | PEMT | 4.42E-05 | 2.300557 | NM_007169 |
H1RNA | H1RNA_X16612_TATA345_chr14_0,0 | 4.87E-05 | 2.22743 |
|
AHSA1 | C14orf3 | 5.03E-05 | 2.224114 | NM_012111 |
CD3D | CD3D | 5.28E-05 | 2.314604 | NM_000732 |
SPK | SPK | 5.29E-05 | 2.196515 | NM_004819 |
PRPF31 | PRPF31 | 5.71E-05 | 2.36207 | NM_015629 |
ZNF133 | ZNF133 | 5.72E-05 | 2.182406 | NM_003434 |
STCH | STCH | 5.84E-05 | 2.2189 | NM_006948 |
CYB561D2 | 101F6 | 6.14E-05 | 2.307171 | NM_007022 |
FLJ21742 | FLJ21742 | 7.27E-05 | 2.307891 | NM_032207 |
ING3 | ING3_0,0 | 7.5E-05 | 2.152426 |
|
CNOT4 | CNOT4 | 9.65E-05 | 2.118939 | NM_013316 |
PET112L | PET112L | 9.98E-05 | 2.284023 | NM_004564 |
ZNF436 | BC056400_0,0 | 0.000105 | 2.123893 |
|
AUTL1 | AUTL1 | 0.00011 | 2.121149 | NM_032852 |
FYCO1 | FYCO1 | 0.000116 | 2.634725 | NM_024513 |
COPS7B | COPS7B | 0.000123 | 2.160383 | NM_022730 |
SEDLP | SEDLP | 0.000131 | 2.117285 | NM_015890 |
HSPC144 | HSPC144 | 0.000132 | 2.227479 | NM_014174 |
HSP105B | HSP105B | 0.000136 | 2.172044 | NM_006644 |
ZFYVE19 | FLJ14840 | 0.000137 | 2.157679 | NM_032850 |
RBL1 | RBL1_0,0 | 0.000147 | 2.0961 |
|
ARL6 | DKFZP434L1123 | 0.000152 | 2.197188 | NM_032146 |
TNFAIP1 | TNFAIP1 | 0.000156 | 2.467579 | NM_021137 |
RPL18A | RPL18A | 0.000157 | 2.100565 | NM_000980 |
C11orf10 | C11orf10 | 0.00016 | 2.083368 | NM_014206 |
BRD8 | BC008076_0,0 | 0.000182 | 2.347046 |
|
VRK3 | LOC51231 | 0.000184 | 2.118904 | NM_016440 |
FNTB | FNTB | 0.000186 | 2.238582 | NM_002028 |
BTRC | BTRC | 0.000194 | 2.06488 | NM_033637 |
PHB | PHB | 0.000196 | 2.141411 | NM_002634 |
GSPT1 | GSPT1 | 0.000197 | 2.099706 | NM_002094 |
KBTBD7 | DKFZP434E2318 | 0.000208 | 2.070793 | NM_032138 |
RBM6 | RBM6 | 0.000243 | 2.01901 | NM_005777 |
ZCCHC8 | DKFZp434E2220 | 0.000248 | 2.135749 | NM_017612 |
PHF6 | MGC14797 | 0.000249 | 1.984857 | NM_032335 |
SEC11L3 | LOC90701 | 0.000252 | 2.091293 | NM_033280 |
CD3EAP | ASE-1 | 0.000255 | 2.174561 | NM_012099 |
IFRD2 | IFRD2 | 0.000255 | 2.047137 | NM_006764 |
QTRTD1 | FLJ12960 | 0.000269 | 2.208297 | NM_024638 |
ZNF317 | BC009367_0,0 | 0.000272 | 2.002955 |
|
ZNF155 | ZNF155_0,0 | 0.000273 | 2.254842 |
|
RNU42B | RNU42B_NR_000013_chr17_0,0 | 0.000276 | 2.013662 |
|
SMAP | IMAGE145052 | 0.000282 | 2.027713 | NM_014267 |
HKE2 | HKE2 | 0.000293 | 1.981017 | NM_014260 |
STARD10 | SDCCAG28 | 0.000302 | 1.961713 | NM_006645 |
CLDND1 | C3orf4 | 0.000309 | 1.975918 | NM_019895 |
RPLP0L | RPLP0L | 0.000311 | 1.982819 | NM_016183 |
SPHK2 | SPHK2 | 0.000318 | 1.948548 | NM_020126 |
ACAD8 | ACAD8_0,0 | 0.00033 | 2.04361 |
|
OSCAR | OSCAR | 0.000351 | 2.27924 | NM_130771 |
SURVIVIN | BIRC5 | 0.000354 | 1.997007 | NM_001168 |
CDK5 | CDK5 | 0.000355 | 1.991764 | NM_004935 |
PSMA1 | PSMA1 | 0.000356 | 1.965821 | NM_002786 |
PEX26 | FLJ20695 | 0.00036 | 2.170953 | NM_017929 |
PCQAP | PCQAP | 0.000365 | 1.952501 | NM_015889 |
ZMAT5 | LOC55954 | 0.000377 | 2.267603 | NM_019103 |
CDC25A | CDC25A_i | 0.000386 | 1.95516 |
|
R3HDM | R3HDM | 0.000388 | 1.952502 | NM_015361 |
SEL1L | SEL1L | 0.000413 | 1.971466 | NM_005065 |
CDC25A | CDC25A | 0.000421 | 1.938726 | NM_001789 |
SLC25A19 | SLC25A19 | 0.000425 | 2.010339 | NM_021734 |
DERL1 | MGC3067 | 0.00043 | 1.966684 | NM_024295 |
BET3 | BET3 | 0.000453 | 1.94045 | NM_014408 |
SLC3A2 | BC001061_0,0 | 0.000493 | 2.302376 |
|
GTF2H4 | GTF2H4 | 0.000501 | 1.932928 | NM_001517 |
PTCRA | PTCRA | 0.000503 | 1.913025 | NM_138296 |
ZNF548 | BC030788_0,0 | 0.000522 | 1.899183 |
|
BUB1B | BUB1B | 0.000559 | 1.926117 | NM_001211 |
TFPT | TFPT | 0.000566 | 2.022086 | NM_013342 |
RNU60 | RNU60_X96660_chr16_0,0 | 0.000583 | 1.911113 |
|
MRPS16 | MRPS16 | 0.000594 | 1.88073 | NM_016065 |
FIS1 | LOC51024 | 0.000598 | 2.009625 | NM_016068 |
ZNF226 | ZNF226_0,0 | 0.000603 | 1.897912 |
|
GMPR2 | LOC51292 | 0.000712 | 1.879694 | NM_016576 |
MRPS11 | MRPS11 | 0.000749 | 1.880522 | NM_022839 |
LGMN | LGMN | 0.00077 | 1.856637 | NM_005606 |
ZNF134 | BC053511_0,0 | 0.000842 | 1.839742 |
|
ZNF24 | ZNF24_0,0 | 0.00085 | 1.872989 |
|
ACP6 | LOC51205 | 0.000858 | 1.861923 | NM_016361 |
SHQ1 | FLJ10539 | 0.000884 | 1.894574 | NM_018130 |
RBL1 | RBL1_i | 0.000906 | 1.912633 |
|
ABCC5 | ABCC5 | 0.000919 | 2.019765 | NM_005688 |
EIF1A | EIF1A | 0.00092 | 1.869566 | NM_001412 |
CBR1 | FLJ10432 | 0.000945 | 1.973468 | NM_019070 |
RNU26 | RNU26_U40580_chr11_0,0 | 0.000951 | 1.952312 |
|
HOM-TES-103 | DKFZP586I2223 | 0.000968 | 1.835571 | NM_015438 |
SPCS2 | KIAA0102_0,0 | 0.000969 | 1.966542 |
|
Table 2. Validation of ChIp-on-chip NOTCH1 target genes
Sequence | ChIP-on-chip P value | Validation | NOTCH1 Val 1744 ChIP Q-PCR fold enrichment |
BUB1B proximal promoter | 5.59 x 10-4 | YES | 2.116 |
BUB3 proximal promoter | 3.12 x 10-8 | YES | 3.574 |
CD3D proximal promoter | 5.28 x 10-5 | YES | 36.41 |
CDC25A proximal promoter | 3.86 x 10-4 | YES | 6.858 |
IFRD2 proximal promoter | 2.55 x 10-4 | YES | 2.751 |
ING3 proximal promoter | 6.53 x 10-7 | YES | 3.32 |
PAFAH2 proximal promoter | 9.78 x10-8 | NO | 0.99 |
PHB proximal promoter | 1.96 x 10-4 | YES | 2.272 |
PRCC proximal promoter | 4.76 x 10-9 | NO | 0.4 |
RBL1 proximal promoter | 1.47 x 10-4 | YES | 2.285 |
TARBP2 proximal promoter | 3.01 x 10-7 | YES | 2.206 |
USP5 proximal promoter | 1.92 x 10-5 | YES | 5.554 |
Table 3. MYC ARACNe neighbor genes in the c-MYC target database
ABCE1 |
C1ORF33 |
C1QBP1 |
CYB5 |
DKC1 |
EEFIG |
EXOSC5 |
HSPCB |
HSPD1 |
IFRD2 |
JTV1 |
NS |
PPAT |
RPL12 |
RPL35 |
RPL7A |
RPS18 |
SCARB1 |
WDR3 |
*www.myccancergene.org/site/mycTargetDB.asp
Supporting Methods
ChIP-on-Chip and Promoter Analysis.
Triplicate NOTCH1 chromatin immunoprecipitations from 5 ´107 to 1 ´ 108 HPB-ALL cells with an antibody recognizing the intracellular TAD domain of NOTCH1 (N1-TAD) kindly provided by Jon Aster were hybridized to HU19k promoter arrays (1, 2). The HU19K genomic array platform contains 13,000 human proximal promoter sequences, typically located between -700 to +200 bp relative to the transcription initiation site, plus sequences encompassing the promoters of all human transcription factor genes up to -3 kb from the transcription initiation site (1, 2). A whole-chip error model (3) that subtracts false positive spots identified in control IgG hybridizations and hybridizations with multiple unrelated antibodies was used to calculate confidence values for each DNA spot on the microarray, to combine data from the replicates of each experiment and to obtain a final average ratio and confidence interval for each promoter region. Raw promoter array scanning data and detailed information about statistical procedures can be found at http://web.wi.mit.edu/young/NOTCH1.For analysis of CSL binding motif frequencies in NOTCH1 target genes, the promoter sequences (-1,000 to +200 bp from the transcription start site) of all genes present in the HU19K arrays were annotated with exact matches to the "TGGGAA" CSL consensus binding site. Differences in the frequency of this CSL DNA binding motif in NOTCH1 target genes (ChIP-on-chip P values <0.001) and in the "background" group containing all other genes on the ChIP-on-chip microarray, were assessed by applying a c2 test to a two-by-two contingency table with the rows containing the number of binding sites and total available sequence length for each group. The P value from this test was 1.10 ´ 10-13.
Gene Expression Analysis.
RNA extracted from experimental duplicates of 7 T-ALL cell lines (ALL-SIL, CCRF-CEM, CUTLL1, DND41, HPB-ALL, KOPTK1 and RPMI8402), treated with GSI (500 nM Compound E) or vehicle only (DMSO) for 24 h, were analyzed with Affymetrix Human U133 Plus 2.0 Arrays. Interarray intensity differences were normalized with Dchip (4). Compound E [(2S)-2-{[(3,5-difluorophenyl)acetyl]amino}-N-[(3S)-1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-1,4-benzodiazepin-3-yl]propanamide] is a cell permeable, potent, selective, non-transition-state, and noncompetitive inhibitor of g-secretase (5, 6). Microarray data are available through the Gene Expression Omnibus repository (accession no. GSE5827).Normalized expression levels with respect to mock-treated controls were calculated for each cell line by dividing expression values of each gene in the array by its mean expression in the two mock-treated samples. Significant changes in gene expression associated with GSI treatment were estimated by using one-sample t test with 13° of freedom. P values are one-tailed based on the hypothesis that NOTCH1 would be an activator, and that therefore, most genes were expected to be down-regulated by GSI treatment.
Functional annotation based on the Gene Ontology classification was performed with the DAVID tool (7).
Gene set enrichment (GSEA) analysis was performed on normalized expression data as described (8) by using signal-to-noise (mclass 0 - mclass 1)/(sclass 0 + sclass 1) to establish the correlation of expression data with DMSO-treated (class 0) and GSI-treated (class 1) groups (9). The rank of genes associated with GSI treatment was tested against NOTCH1 ChIP-on-chip target genes and genes associated with forced expression of c-MYC in T cell precursors (10). Genes associated with enforced expression of MYC in murine T cell lymphoblasts were identified by supervised analysis with D-chip default settings on public microarray data published by Marikovic et al. (10). Correspondence between mouse genes and human Affymetrix identifiers was established by using NetAffx Analysis Center tools supported in the Affymetrix web site.
An ARACNe mutual information network (11) was built based on a large panel of data from primary human T-ALL samples (n = 99), cell lines (n = 3), and normal thymus (n = 1) using Human U133A GeneChip data (12) after GC-RMA array normalization. A metagene entry, constructed to be proportional to the concentration of activated NOTCH1 protein in the nucleus, was constructed by using stepwise regression from quantitative measurements of ICN1 protein level in T-ALL cell lines. The linear model considers quantified Western blot of activated NOTCH1 as the response variable and expression level of other genes as the explanatory variables. To reduce the variable search space and possible overfitting, only genes highly correlated with activated NOTCH1 were considered as candidates. The model was developed in stages by adding the best explanatory variable one by one until the model converged. The identified explanatory variables are the expression levels of COX7A2L, TRO, PPP2R4, PARP8, HSPD1, and E2F8. This computed metagene was integrated into the microarray gene expression profile measurements and used to build an ARACNe network for the NOTCH1 signaling hub (13). Analysis of the NOTCH1 metagene and c-MYC ARACNe neighbors identified 12 genes, most of them involved in protein biosynthesis, shared between these two hubs. In addition, conditional mutual information of the two hubs (conditioned on their common neighbors) does not increase compared to the original pairwise mutual information, indicating a direct close relationship between the NOTCH1 metagene and c-MYC, consistent with a feed-forward-loop regulatory motif model.
Quantitative ChIP analysis
. Relative quantitation by real time PCR of c-MYC promoter sequences (-1807 to -1597 from the transcription initiation site) were normalized to beta actin levels in chromatin immunoprecipitates performed with antibodies against the transactivation domain of NOTCH1 (N1-TAD), the activated gamma-secretase cleaved form of NOTCH1 (Val-1744; Cell Signaling) and IgG (used as negative control). Similarly, 12 randomly selected NOTCH1 target genes identified by ChIP-on-chip (Table 2) were analyzed in independent chromatin immunoprecipitates performed with the NOTCH1 Val-1744 antibody.Quantitative Real-Time PCR.
Total RNA from T-ALL cell lines was extracted with RNAqueous kit (Ambion) according to the manufacturer's instructions. cDNA was generated with the ThermoScript RT-PCR system (Invitrogen) and analyzed by quantitative real-time PCR (SYBR Green RT-PCR Core Reagents kit and the 7300 Real-Time PCR System, both from Applied Biosystems). Relative expression levels were based on GAPDH levels used as reference controls. Primer sequences are available upon request.Cell Cycle Analysis.
Cell cycle distribution was performed by assessment of DNA content using PI staining as described (14).1. Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. (2004) Science 303:1378-1381.
2. Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. (2005) Proc Natl Acad Sci USA 102:4459-4464.
3. Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. (2002) Science 298:799-804.
4. Li C, Wong WH (2001) Proc Natl Acad Sci USA 98:31-36.
5. Seiffert D, Bradley JD, Rominger CM, Rominger DH, Yang F, Meredith JE, Jr, Wang Q, Roach AH, Thompson LA, Spitz SM, et al. (2000) J Biol Chem 275:34086-34091.
6. Weng AP, Ferrando AA, Lee W, Morris JP t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC (2004) Science 306:269-271.
7. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) Genome Biol 4:P3.
8. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Proc Natl Acad Sci USA 102:15545-15550.
9. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. (1999) Science 286:531-537.
10. Marinkovic D, Marinkovic T, Kokai E, Barth T, Moller P, Wirth T (2004) Nucleic Acids Res 32:5368-5378.
11. Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005) Nat Genet 37:382-390.
12. Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, Baruchel A, Toribio ML, Sigaux F (2005) Blood 106:274-286.
13. Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A (2006) BMC Bioinformat 7(Suppl 1):S1--S7.
14. Look AT, Melvin SL, Brown LK, Dockter ME, Roberson PK, Murphy SB (1984) J Clin Invest 73:1617-1628.