Supplementary material for Kapadia et al. (1999) Proc. Natl. Acad. Sci. USA 96 (21), 12073-12078.
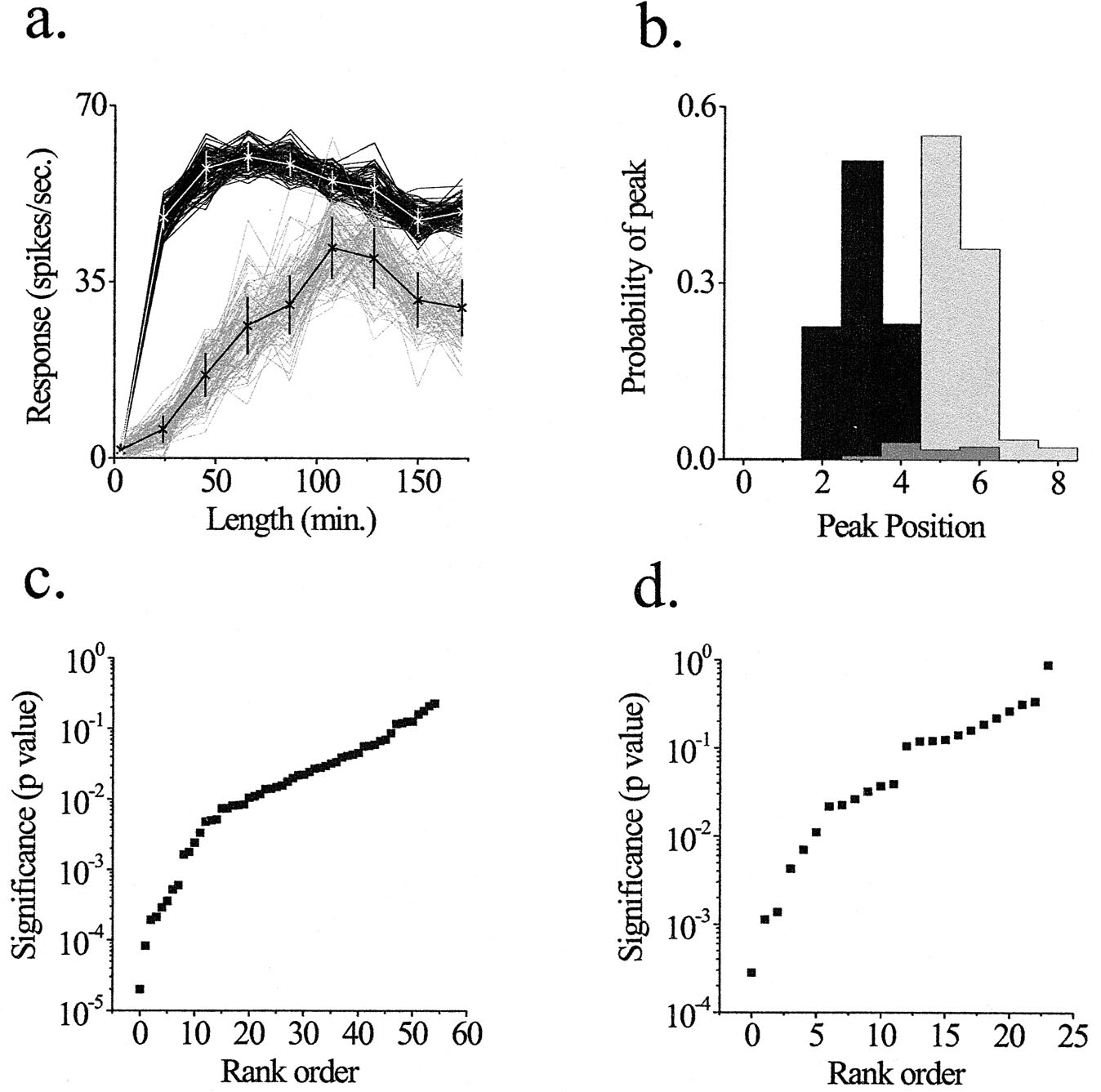
Fig. 6.
Monte Carlo simulations. Monte Carlo simulations were used to test the statistical significance of changes in spatial summation on a cell-by-cell basis. (a) Construction of simulated tuning curves. For each tuning curve measured experimentally, we generated 10,000 simulated tuning curves in which each point was randomly assigned a value from a normal distribution with a mean and bandwidth taken from the mean and the standard error of the biological data at that point. The first 100 simulations are shown for high-contrast (black curves) and low-contrast (light gray curves) conditions in one cell. The physiological data are plotted over the simulated curves with standard errors. (b) Probability distributions. The largest response from each simulated tuning curve is defined as its peak, and the probability of the peak occurring at each position is calculated from the 10,000 simulations. The distribution of peak positions is plotted for the two contrast conditions shown in a. The probability that the two distributions share the same peak is calculated from the overlap of the two distributions (dark gray). For this cell, p = 0.026. (c) Significance values for high- vs. low-contrast comparisons. The distribution of p values is plotted as a function of their rank order. Of the 55 cells, 41 (75%) reached statistical significance at the level of p < 0.05. (d) Significance values for high-contrast vs. high-contrast/noisy background comparisons. Of the 24 cells, 12 (50%) reached statistical significance.