Lam et al. 10.1073/pnas.0611081104. |
Fig. 5. A map showing the location of our sampling station (in red) in the Black Sea, and the generalized surface circulation (in blue) within the basin (1).

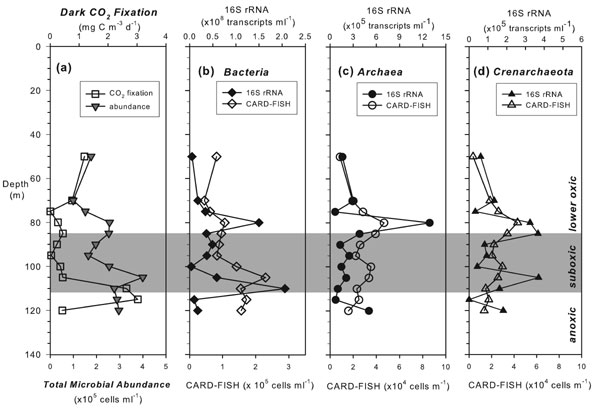
Fig. 6. Vertical profiles of dark inorganic carbon fixation rates and total microbial abundance measured by flow cytometry (a); and the CARD-FISH cell counts and 16S rRNA transcript levels of Bacteria (b), Archaea (c), and Crenarchaeota (d). The shown CARD-FISH counts are mean values with standard deviations of 6%, 14%, and 9% for Bacteria, Archaea, and Crenarchaeota, respectively.
Fig. 7. A consensus phylogenetic tree of metabolically active Archaea based on 16S rRNA obtained from reverse transcription PCR (RT-PCR) and subsequent clone library construction. The branching pattern has been verified by maximum likelihood, >50% bootstrapped maximum parsimony (1,000´) and distance matrix (1,000´) methods. Only representative operational taxonomic units (<97% sequence identity cutoff) are shown. Blue sequences were retrieved from 80 m, green from 100 m, and red from 105 m. Only sequences >1,000 bp are included in the analyses, whereas AY360478 and AY360479 (both <600 bp) were added to the consensus tree afterwards using parsimony method in ARB.
Fig. 8. Temporal variations of dissolved oxygen, temperature-salinity ratio, and transmission over a period of ≈22 h, and a vertical profile of amoA transcript abundance from g-proteobacterial AOB, all plotted against density (s t) equivalent to 80-120 m.

Fig. 9. A proposed revision to the marine nitrogen cycle with direct nitrification-anammox coupling in the suboxic zone. It illustrates the fact that nitrification and anammox, previously considered as clearly oxic and anoxic processes respectively, can actually cooccur under microaerobic conditions. Nitrification in the suboxic zone provides a direct source of nitrite for anammox. It acts as a short circuit channeling regenerated NH4+ to direct N loss. If this coupling, as observed in the Black Sea, also occurs in oceanic OMZs, then the relative contribution directly from upwelled nitrate to marine N loss will be less than previously considered. Meanwhile, the occurrence of the classic heterotrophic denitrification (indicated as '?') as a source of direct nitrogen loss is undetectable in the Black Sea suboxic waters, as well as in the Peruvian-Chilean (2, 3) and Benguelan (4) OMZs. (Modified from ref. 5.)
1. Sorokin YI (2002) The Black Sea: Ecology and Oceanography (Backhuys Publishers, Leiden, The Netherlands).
2. Hamersley MR, Lavik G, Woebken D, Rattray JE, Lam P, Hopmans EC, Sinninghe Damsté JS, Krüger S, Graco M, Gutiérrez D, Kuypers MMM Limnol Oceanogr, in press.
3. Thamdrup B, Dalsgaard T, Jensen MM, Ulloa O, Farías L, Escribano R (2006) Limnol Oceanogr 51:2145-2156.
4. Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, Jorgensen BB, Jetten MSM (2005) Proc Natl Acad Sci 102:6478-6483.
5. Arrigo KR (2005) Nature 437:349-355.
Table 1. Gene copy ratios of crenarchaeal ammonia monooxygenase subunit A (amoA) to 16S rRNA genes, and of crenarchaeal to total bacterial (AOB) amoA genes detected across the Black Sea oxic, suboxic, and anoxic zones
Type | Depth, m | Crenarchaeal amoA:16S rRNA gene copy ratios | Crenarchaeal:AOB amoA gene copy ratios |
Oxic | 50 | 0.33 | 4.6 |
Oxic | 63 | 0.64 | 30.0 |
Oxic | 85 | 0.68 | 44.1 |
Suboxic | 100 | 1.2 | 4.4 |
Suboxic | 105 | 2.8 | 40.6 |
Suboxic/ anoxic | 110 | 0.04 | 0.4 |
Anoxic | 120 | 0.04 | 0.4 |
Anoxic | 147 | 0.01 | 0.6 |
SI Materials and Methods
Nucleic Acids Extraction, PCR, RT-PCR, Cloning, and Phylogenetic Analyses.
DNA samples were collected by large-volume in situ filtration on to cellulose acetate membrane filters (0.2 mm pore-size) and extracted as described in (1). Crenarchaeal amoA genes were PCR-amplified (2), cloned with TOPO TA Cloning Kit (Invitrogen) and sequenced with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturers' instructions. Phylogenetic analyses were performed with the ARB software package (3). RNA samples were collected by filtering 5-10 L of seawater into Sterivex filters (0.22 mm pore-size, Millipore) and immediately frozen at -80°C. Cell lyses were done following (4) with an additional 10 units ml-1 of RNase inhibitor (SUPERaseIn, Ambion). RNA was subsequently purified from cell lysates using the Qiagen RNA/DNA kit (Qiagen) and DNase-treated (TURBO DNA-free, Ambion). RNA quantities and integrity were checked with the Agilent 2100 Bioanalyzer (Agilent Technologies). Reverse transcription (RT) was conducted with the Supercript III First-Strand Synthesis Kit (Invitrogen). Gene-specific primers were used in the RT: the universal reverse primer 1392R (5) targeting 16S rRNA, and the antinsense primers targeting crenarchaeal and bacterial amoA genes as described below. An RNaseH treatment was conducted to remove residual RNA. Selected cDNA samples were PCR-amplified, cloned and sequenced for phylogenetic analyses. The primer pairs used were: A20f (6)/1392R (5), Arch-amoAF/R (2), amoA-1F/2R (7), amoA3F/amoB4R (8), targeting archaeal 16S rRNA, amoA from Crenarchaeota, bAOB and gAOB respectively. The same cycling parameters were applied as in the original primer designs.Quantitative PCR and RT-qPCR.
The TaqMan fluorogenic PCR method (9) (TaqMan Universal Master Mix, Applied Biosystems) was used to quantify the 16S rRNA (cDNA from the RT) of Eubacteria (10), Archaea (11), anammox bacteria (only DNA) (12) and Crenarchaeota. A new primer and probe set was developed for Crenarchaeota 16S rRNA using the Primer Express v.2.0 software (Applied Biosystems) based on sequences retrieved in the GenBank (13): Cren334F (5'- AGA TGG GTA CTG AGA CAC GGA C-3'), Cren554R (5'- CTG TAG GCC CAA TAA TCA TCC T-3') and probe Cren519 (5'-TTA CCG CGG CGG CTG ACA C-3'). The specificity of their sequences was verified with the ARB software package, BLAST (14), and the Probe Match tool from the Ribosomal Database Project II (15). The cycling parameters were 2 min at 50°C, 10 min at 95°C followed by 40 cycles of 15s at 95 and 1 min at 60°C. The cDNA generated by the RTs with the universal 1392R primer was used for different 16S rRNA quantification. To quantify amoA genes and cDNA, the SYBR Green assay (Power SyBr Green, Applied Biosystems) was used with the same primer sets and cycling parameters as described for regular RT-PCR, except that 50 cycles were performed and the signals were recorded at a higher than annealing temperature (78°C) to avoid primer-dimers. Specificity of the PCR products was checked with postreal-time-PCR melt-curves and agarose gel electrophoreses. DNA extracts from pure Escherichia coli, Nitrosomonas eutropha, Nitrosospira multiformis and Nitrosococcus oceani cultures, anammox bioreactor enrichment cultures, were used as our Eubacteria, bAOB, gAOB and anammox bacteria (RT)-qPCR standards. Plasmids obtained from cloned and sequenced marine Archaea were used as the (RT)-qPCR standards for archaeal and crenarchaeal 16S rRNA, as well as crenarchaeal putative amoA genes. All (RT-)qPCR reactions were run as triplicates from the same DNA or cDNA sample on an iQ5 Real-Time PCR System (Bio-Rad Laboratories).1. Zhou JM, Bruns A, Tiedje JM (1996) Appl Envir Microbiol 62:316-322.
2. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Proc Natl Acad Sci USA 102:14683-14688.
3. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, et al. (2004) Nucleic Acids Res 32:1363-1371.
4. Somerville CC, Knight IT, Straube WL, Colwell RR (1989) Applied Environ Microbiol 55:548-554.
5. Stahl DA, Amann R (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E, Goodfellow M (Wiley, Chichester, England), pp 205-248.
6. Massana R, Murray A, Preston C, DeLong E (1997) Appl Environ Microbiol 63:50-56.
7. Rotthauwe J-H, Witzel K-P, Liesack W (1997) Applied Environ Microbiol 63:4704-4712.
8. Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops H-P, Wagner M (2000) Applied Environ Microbio 66:5368-5382.
9. Livak KJ, Flood SJA, Marmaro J, Giusti W, Deetz K (1995) PCR Methods Appl 4:357-362.
10. Nadkarni MA, Martin FE, Jacques NA, Hunter N (2002) Microbiology 148:257-266.
11. Takai K, Horikoshi K (2000) Appl Environ Microbiol 66:5066-5072.
12. Hamersley MR, Lavik G, Woebken D, Rattray JE, Lam P, Hopmans EC, Sinninghe Damsté JS, Krüger S, Graco M, Gutiérrez D, Kuypers MMM (in review) Limnol. Oceanogr.
13. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2006) Nucleic Acids Res 34:D16-D20.
14. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25:3389-3402.
15. Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) Nucleic Acids Res 31:442-443.