Kandoth et al. 10.1073/pnas.0700344104. |

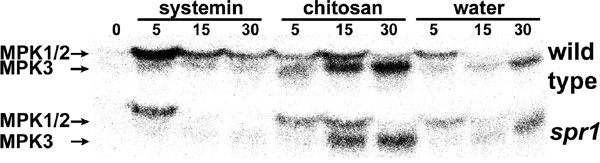
Fig. 6. MPK1/2 activation by systemin is SPR1-dependent. Fourteen-day-old wild-type tomato seedlings (var. Castlemart) and spr1 mutant plants (in Castlemart background) were excised at the base of the stem and either placed in water or a solution containing systemin (25 nM) or chitosan (125 mg/ml) or left untreated (0). At the times indicated (minutes), plants were frozen in liquid nitrogen, and leaf extracts were analyzed for MAPK activity using an in-gel kinase assay. The higher molecular mass band (48 kDa) represents LeMPK1/2 activity and the lower molecular mass band (44 kDa) represents LeMPK3. The experiment is representative of three similar experiments.

Fig. 7. (A) Cosilencing of MPK1 and MPK2 attenuates ethylene synthesis. 35S::prosys plants were infiltrated with either pTRV-MPK1/2 (open bars) or pTRV-GFP (Control; solid bars). Four weeks later, leaves were excised at the petiole and incubated for 2 h in a sealed Petri dish. At 1 and 2 hr after excision, ethylene concentrations were measured by gas chromatography. Bars represent mean and SEM from two independent experiments. Ethylene production increased with time (P < 0.001), but was significantly decreased in pTRV-MPK1/2 plants (P < 0.03) (repeated measures analysis of variance using PROC MIXED in SAS 9.3.1). (B) The excised leaves were then analyzed for PI-II protein content by a radial immunodiffusion assay to demonstrate the effect of VIGS. PI-II levels in pTRV-MPK1/2-infiltrated plants (open bar) were expressed as the percentage of mean levels in control plants (solid bar).
SI Materials and Methods
VIGS Constructs.
Semiquantitative RT-PCR (sqRT-PCR)
. RNA isolation from leaves and reverse transcription were described (4). PCR cycle conditions were 95°C for 1 min, followed by 35 cycles of 15 s at 95°C, 30 s at 56°C, and 45 s at 72°C, and a final extension of 7 min at 72°C. Five-microliter aliquots were collected at two or three cycle intervals as indicated in the text. PCR products were separated by agarose gel electrophoresis followed by ethidium bromide staining. "No-RT" controls demonstrated the absence of genomic DNA contamination. Actin transcript levels were used as internal controls for normalization. The following sets of primers were used for sqRT-PCR to analyze silenced plants. They amplified sequences outside the target sequence in the pTRV2 vector. For analysis of MPK1/2-cosilenced plants, primers which amplify parts of the 3'UTR of MPK1 (forward GCTGACAGATTGTTGCAGGT; reverse TCCACCCCATAAAGATACATCA; 172 bp) and MPK2 (forward TACTCGCTCGTTTGCTGTTG; reverse TTGGAGTACAGGAAAACAATGG; 170 bp) were used. To assess transcript levels of MPKs in experiments that target only one specific MPK, the following primers were used. MPK1: forward TGCACCTCCGGTCAACAA; reverse GGCAGTGCTCCTCAGATAAA; MPK2: forward GGAGAAAAGAGGGAAAATTTGA; reverse GGCAATGTTCTTCTGATAAT; MPK3 forward CTAAATTTCTATCAATAATGGTTGATGC; reverse GCGGAGGAATCACATCTCTT. To determine the expression levels of wound response genes by sqRT-PCR, the following primers were used. LeAOS2 (GenBank accession no. AF230371): forward GCAACGAAGGATCCGAAAAT, reverse ACTGGCCGATAGTGACAGTG, 344 bp long; LeLoxD (GenBank accession no. U37840): forward CCGTGGTTGACACATTATCG, reverse ACAGCAGTCCGCCCTATTTA, 344 bp long; LeAOC (GenBank accession no. AJ272026): forward GCACCTCAACAGATTCAACTAACACTG, reverse GTAATTTTCAGTGCGGCCCCTTCC, 533 bp long; LePI-I (GenBank accession no. K03290): forward TGAAACTCTCATGGCACGAA, reverse TTTTGACATATTGTGGCTGCTT, 331 bp long; LePI-II (GenBank accession no. K03291): forward CCCACGTTCAGAAGGAAGTC, reverse TTTTGGGCAATCCAGAAGAT, 361 bp long; LeACTIN (GenBank accession no. U60478): forward ATGACTCAAATCATGTTTGAGACCTTC, reverse ACCTTAATCTTCATGCTGCTTGGAGC, 631 bp long. The band intensities in the linear range of amplification were quantified using ImageQuant version 5.2 software (Molecular Dynamics, Sunnyvale, CA). For this, the band intensities were first normalized with respect to actin mRNA levels in control plants. The normalized band intensity in silenced plants was expressed as percentage of mRNA levels in control plants.
Jasmonic Acid (JA) Analysis: Gas Chromatography-Mass Spectroscopy (GC-MS).
Volatiles eluted from a SuperQ matrix (Alltech Associates, State College, PA) were analyzed by GC-MS using electron impact ionization (EI) in selective ion mode. Briefly, an HP 5890 gas chromatograph equipped with a split/splitless injector (splitless mode, injection volume 1 ml) was interfaced to an VG-70S magnet sector mass spectrometer (Waters Corp, Milford, MA). Compounds were separated on an Restek RTx-5 (30 m in length, 0.25-mm ID, 0.2-mm film) column preheated to 80°C. After injection, the temperature was increased at 10°C/min to 130°C, then 3°C/min to 180°C, and finally 10°C/min to 300°C, and held at 300°C for 10min. Helium was used as the carrier gas at 10 psi. Specific EI conditions were 70 eV, selection monitoring at 4,000 resolution. Retention time for the JA (224.1412) transisomer was 18.35 min, for the cis-isomer 19.35 min, for the dhJA (226.1569) transisomer 18.44 min, and 19.36 min for the cis-isomer.
The internal standard dhJA was prepared from dihydro methyl jasmonate (a kind gift from Bedoukian Research, Danbury, CT) by alkaline hydrolysis according to Dathe et al. (5).
Ethylene Analysis.
Leaves of 4-week old pTRV-MPK1/2-infiltrated 35S::prosys plants (var. MicroTom) or pTRV-GFP-infiltrated 35S::prosys control plants were excised at the petiole. Leaflets were weighed (»0.5 g for each sample) and then incubated for 2 hr at 27°C under bright light in a sealed Petri dish featuring a septum. Five-hundred-microliter air samples were collected from the Petri dish with an airtight syringe at 1 and 2 hr after excision and immediately injected into a gas chromatograph [Shimadzu GC-8A, column: Porapak T 80/100 (Altech), 180 ´0.31 ´ 0.085 cm; N2 Carrier gas] equipped with a flame ionization detector. Ethylene eluted with a retention time of 0.91 min which was verified with an authentic standard of ethylene (National Welders, Charlotte, NC).
1. Liu Y, Schiff M, Dinesh-Kumar SP (2002) Plant J 31:777-786.
2. Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Plant J 39:734-746.
3. Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Proc Natl Acad Sci USA 94:2122-2127.
4. Higgins R, Lockwood T, Holley S, Yalamanchili R, Stratmann J (2007) Planta 225:1535-1546.
5. Dathe W, Rönsch H, Preiss A, Schade W, Sembdner G, Schreiber K (1981) Planta 153:530-535.