Korczynska et al. 10.1073/pnas.0701809104. |

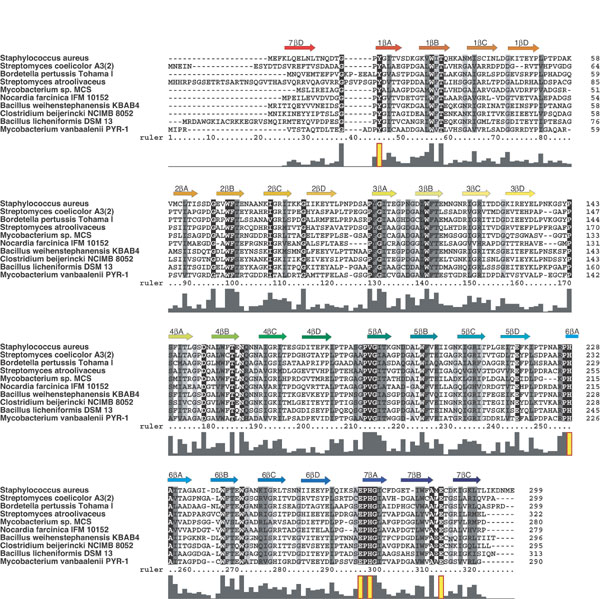
Fig. 4. Sequence conservation among Vgb orthologues. The sequence of Vgb from S. aureus is aligned against eight other Vgbs. Conservation of residues is highlighted as a gradient from black (strictly conserved) to light gray (weakly conserved). Secondary structure assignments are shown above the alignment, and a bar graph indicating degree of conservation is shown below. Residues important for Vgb function are highlighted (yellow) in the conservation bar graph.

Fig. 5. Repeating motif within the sequence of Vgb. (A) Structure-based sequence alignment of the blades of Vgb. The b-strands are indicated above the sequence. Conservation of residues is highlighted as a gradient from black (strictly conserved) to light gray (weakly conserved). Residues important for Vgb function are boxed. The consensus sequence repeat is indicated below the alignment. (B) Comparison between repeating motifs observed in proteins with a b-propeller architecture. In addition to Vgb, schematic diagrams are shown for WD40, Kelch, YWTD, PQQ, and AxSPD consensus repeats.

Fig. 6. Crystal packing contacts in VgbWT crystals. (A) Arrangement of the four Vgb molecules in the asymmetric unit. Highlighted in purple are b-strands D and A of blades 1 and 2, respectively, and the connecting loop, for two of the four Vgb molecules. This loop binds to a depression on the top face of a neighboring molecule, thereby obstructing the quinupristin binding site. (B) Close-up view of the packing interaction. Also shown in green is the loop replacement introduced (51-56 PLPTPD to ELPNKG) to disfavor this particular packing interaction and allow for structure determination of the substrate-bound complex. (C) Schematic view of quinupristin-bound Vgb in an identical orientation as B, illustrating how the packing interaction would interfere with quinupristin binding. It is intriguing to note that the proline moiety of quinupristin interacts similarly with Vgb as the proline present in the loop.

Fig. 7. Binding of dalfopristin to Vgb. (A) Diagram illustrating the arrangement of Vgb molecules present in the VgbI+L•quin•Mg2+ crystal form. Shown in sticks are dalfopristin (orange) and quinupristin (purple). (B) Diagram displaying the surfaces for molecules within 5 Å of dalfopristin. Shown in green and pale yellow are the surfaces for two Vgb molecules, which wedge dalfpristin, and in purple is the surface originating from a nearby quinupristin molecule. (C) 2Fo - Fc density map for dalfopristin contoured at 1s.
Table 3. Data collection and refinement statistics
VgBSe-Met | VgBWT | VgBI+L•quin•Mg2+ | |||
Space group | C2 | P212121 | P212121 | ||
Unit cell parameters, Å, ˚ | a = 93.0, b = 34.8, c = 86.6, b = 117.7 | a = 73.5, b = 79.2, c = 188.2 | a = 71.0, b = 93.7, c = 95.3.2 | ||
Wavelength, Å | 0.9000 | 0.9798 | 1.0600 | 1.0999 | 1.5418 |
Resolution range, Å | 50-1.65 (1.75-1.65) | 50-1.75 (1.81-1.75) | 50-1.90 (1.97-1.90) | 50-1.9 (1.97-1.90) | 50-2.8 (2.8-2.99) |
Completeness, % | 96.7 (78.2) | 94.3 (65.4) | 93.8 (65.1) | 98.9 (93.9) | 91.1 (94.1) |
Redundancy | 7.5 (6.0) | 7.2 (6.1) | 3.6 (3.1) | 4.5 (3.1) | 4.1 (3.8) |
I / s(I) | 18.5 (3.7) | 21.5 (6.6) | 21.6 (9.9) | 19.4 (3.5) | 6.0 (3.8) |
No. reflections | 29,320 | 22,397 | 17,759 | 86,401 | 14,813 |
R sym,* % | 3.8 (38.3) | 3.5 (22.8) | 2.4 (8.9) | 8.1 (46.1) | 13.6 (31.0) |
R factor/Rfree, % | 15.7/20.5 | 14.4/20.5 | 26.3/31.9 | ||
No. atoms | |||||
Protein | 2,312 | 9,337 | 4,548 | ||
Synercid | - | - | 242 | ||
Cl- ions | 1 | 5 | - | ||
Mg2+ ions | - | - | 4 | ||
Water | 210 | 1,083 | 64 | ||
Bond length, Å | 0.026 | 0.022 | 0.007 | ||
Bond angle, ˚ | 2.4 | 1.9 | 1.4 | ||
B factors, Å2 | |||||
Protein | 21.6 | 23.8 | 20.1 | ||
Synercid | - | - | 26.3 | ||
Cl- ions | 10.8 | 25.5 | - | ||
Mg2+ ions | - | - | 9.4 | ||
Water | 29.8 | 38.3 | 19.2 | ||
Data in parentheses represent the outermost shell.