
Craigo et al. 10.1073/pnas.0706449104. |

Fig. 4. Comparison of longitudinal population divergence during experimental infection with EIAVPV. Genetic distances of the deduced amino acid sequences were calculated by using Distance software with Kimura correction (GCG Wisconsin Package of software). Distances were calculated from aligned gp90 regions longitudinal isolates. Calculated distances were plotted as the divergence from the inoculating strain, EIAVPV. The days after infection are indicated below the x axis in blue. (-), mean divergence; I, II, III, IV, V, VI, fever episodes; Inapparent, inapparent stage of disease; È, population subclones chosen for variant challenge strain development.
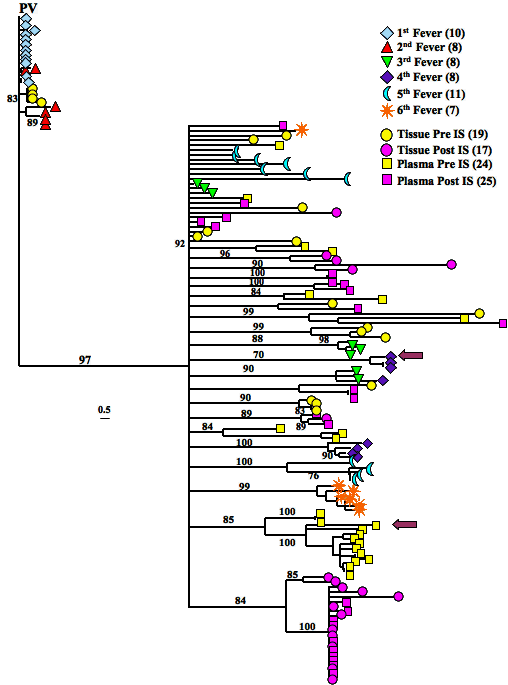
Fig. 5. Evolutionary analyses of the longitudinal isolates derived from experimental infection with EIAVPV. A phylogenetic tree of aligned plasma and tissue immune suppression (inapparent stage) and febrile deduced amino acid sequences (gp90 regions) were constructed by the neighbor-joining method from Kimura-corrected distances with the optimality criterion set to distance. The tree was rooted to EIAVPV. Bootstrap values were determined over 1,000 iterations and are indicated at the nodes of the branches. Branch lengths are proportional to the distance existing between the sequences. IS, immune suppression; Å, population subclones chosen for variant challenge strain development.
Fig. 6. Comparison of deduced amino acid gp90 variable-region sequences from EIAV isolates chosen for variant challenge strain construction. The region of env gene coding for the surface glycoprotein was sequenced from EIAVPV stock and from plasma viral RNA collected during febrile episodes of an EIAVPV-infected pony. Deduced amino acid sequences were aligned and compared with EIAVD9. Only the amino acid residues different from EIAVPV/D9 are reported on the alignment. Dots (·) indicate residues identical to the EIAVPV/D9 sequence; dashes (-) indicate amino acid deletion; previously described variable regions V1-V8 are boxed and shaded yellow; * indicate areas of increased immunological interest.

Fig. 7. Neutralization profiles of proviral variant strains with serum from animals experimentally infected with EV0, EV6, or EV13. Variant proviral strains were tested for in vivo pathogenecity through experimental infections of ponies. At 6 months after infection, serum from two representative animals (labeled A and B) was tested in our standard neutralization assay. The mean reciprocal dilutions of serum that neutralized 50% of input EV0, EV6, or EV13 as measured in an infectious center assay are presented. The line (─) denotes the cut off (≥25) value for valid 50% neutralization titers.

Fig. 8. Clinical and virological profiles of EV0 challenge strain vaccine trial group. (A) Eight EIAV-naïve ponies were vaccinated with 103 TCID50 EIAVD9 i.m. (#Vax#). Rectal temperature (red line, right y axis) and platelet counts (blue dashed line, first left y axis) were followed daily for up to 370 days (x axis) after the first vaccine dose. Quantification of the virus load (¨, second left y axis) was performed on viral RNA extracted from plasma at periodic time points before and after virulent virus challenge 7 months after the first vaccination with 103 TCID50 EV0, I.M (#Challenge). (B) Six EIAV-naïve ponies were also challenged with 103 TCID50 EV0, i.m. (#Challenge). A 14-day period of immune suppression with 0.1mg/kg dexamethasone is indicated by a pink shaded box. Febrile episodes were defined by a achieving a combination of two to three features such as: rectal temperature >39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Diagnostic results of viral strain for each vaccinated animal are indicated in each respective panel above the date of analysis by either a V (vaccine strain) or a C (challenge strain): lower case letters represent the minor population if both vaccine and challenge strain viruses were detected. Pink box, animal killed due to severe disease; gray box, animal killed as endpoint of study.

Fig. 9. Clinical and virological profiles of EV6 challenge strain vaccine trial group. (A) Eight EIAV-naïve ponies were vaccinated with 103 TCID50 EIAVD9 I.M. (#Vax#). Rectal temperature (red line, right y axis) and platelet counts (blue dashed line, first left y axis) were followed daily for up to 370 days (x axis) after the first vaccine dose. Quantification of the virus load (¨, second left y axis) was performed on viral RNA extracted from plasma at periodic time points before and after virulent virus challenge 7 months after the first vaccination with 103 TCID50 EV6, I.M (#Challenge). (B) Six EIAV-naïve ponies were also challenged with 103 TCID50 EV6, I.M (#Challenge). A 14-day period of immune suppression with 0.1mg/kg dexamethasone is indicated by a pink shaded box. Febrile episodes were defined by a achieving a combination of two to three features such as: rectal temperature >39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Diagnostic results of viral strain for each vaccinated animal are indicated in each respective panel above the date of analysis by either a V (vaccine strain) or a C (challenge strain): lower-case letters represent the minor population if both vaccine and challenge strain viruses were detected. Pink box, animal killed due to severe disease;gray box, animal killed as endpoint of study.
Fig. 10. Clinical and virological profiles of EV13 challenge strain vaccine trial group. (A) Eight EIAV-naïve ponies were vaccinated with 103 TCID50 EIAVD9 I.M. (#Vax#). Rectal temperature (red line, right y axis) and platelet counts (blue dashed line, first left y axis) were followed daily for up to 370 days (x axis) after the first vaccine dose. Quantification of the virus load (¨, second left y axis) was performed on viral RNA extracted from plasma at periodic time points prior to and after virulent virus challenge 7 months after the first vaccination with 103 TCID50 EV13, I.M (#Challenge). (B) Six EIAV-naïve ponies were also challenged with 103 TCID50 EV13, I.M (#Challenge). Febrile episodes were defined by a achieving a combination of two to three features such as: rectal temperature >39°C in conjunction with thrombocytopenia (platelet decrease of ≥70,000/µl of whole blood), EIAV viral load ≥105 as well as other clinical signs of EIA. Diagnostic results of viral strain for each vaccinated animal are indicated in each respective panel above the date of analysis by either a V (vaccine strain) or a C (challenge strain): lower-case letters represent the minor population if both vaccine and challenge strain viruses were detected. Pink box, animal killed due to severe disease; gray box, animal killked as endpoint of study.

Fig. 11. Delayed-type hypersensitivity analysis of EIAVD9 vaccinated, immune suppressed animals. Immune status of dexamethasone-treated animals was monitored through a delayed-type hypersensitivity assay. DTH responses were determined by subcutaneous injection of 50 µg of PHA in 1 ml of saline and 1 ml of saline alone into the necks of the horses. Twenty-four hours after injection, the diameter of the reaction was measured using constant tension calipers. The control reaction (saline alone) was divided into the PHA reaction to yield the DTH index (y axis). Pre-IS, pre-immune suppression; IS, immune suppression.
Table 3. Trial subjects
Challenge group | Pony number | Age, years | Gender |
EV0 | A27 | 7 | F |
C27 | 5 | F | |
C28 | 5 | F | |
C34 | 5 | F | |
E31 | 3 | M | |
E33 | 3 | M | |
E34 | 3 | F | |
E35 | 3 | F | |
Controls | D37 | 4 | M |
F40 | 2 | F | |
A23 | 6 | M | |
9955 | 6 | M | |
9913 | 7 | F | |
9961 | 7 | M | |
EV6 | 558 | 12 | M |
A32 | 7 | F | |
A33 | 7 | M | |
C29 | 5 | M | |
C33 | 5 | F | |
C36 | 5 | M | |
D31 | 4 | M | |
D44 | 4 | M | |
Controls | F35 | 2 | F |
598 | 13 | M | |
10 | 10 | M | |
50 | 10 | F | |
B32 | 4 | F | |
956 | 6 | F | |
EV13 | 886 | 9 | F |
B22 | 6 | M | |
B27 | 6 | M | |
D34 | 4 | M | |
D38 | 4 | M | |
D39 | 4 | M | |
D40 | 4 | M | |
E30 | 3 | M | |
(Controls) | D33 | 4 | M |
F38 | 2 | M | |
A26 | 6 | M | |
9959 | 7 | M | |
9 | 8 | M | |
B23 | 5 | M |
Table 4. Trial outcome
Challenge group | Pony number | Acute disease (DPC) | Chronic disease (DPC) | Viral strain detected |
EV0 | A27 | NA | NA | V |
C27 | NA | NA | V | |
C28 | 49 | 72, 75, 83* | V/c | |
C34 | NA | NA | V | |
E31 | NA | NA | V | |
E33 | NA | NA | V | |
E34 | NA | NA | V/c | |
E35 | NA | NA | V | |
Controls | D37 | 14 | NA | C |
F40 | 14 | NA | C | |
A23 | 14 | NA | C | |
9955 | 28 | 106,112,174, 177,178* | C | |
9913 | 43 | NA | C | |
9961 | 12 | NA | C | |
EV6 | 558 | 116 | NA | C/v |
A32 | NA | NA | C | |
A33 | 5 | 27, 87 | C | |
C29 | NA | NA | V | |
C33 | NA | NA | V | |
C36 | 48 | 56, 67, 71, 72* | C | |
D31 | NA | NA | C | |
D44 | NA | NA | V | |
Controls | F35 | 144 | NA | C |
598 | 14 | 66 | C | |
10 | 16 | 49,58,65* | C | |
50 | 17 | 63, 100, 124, 126, 130* | C | |
B32 | 16 | NA | C | |
956 | 19 | NA | C | |
EV13 | 886 | 71 | 101, 102 | C |
B22 | 14 | 34, 44, 48, 51 | C | |
B27 | 36 | 52, 54 | C | |
D34 | NA | NA | C | |
D38 | 86 | NA | C | |
D39 | 17 | 83, 84, 104 | C | |
D40 | NA | NA | C/v | |
E30 | NA | NA | C/v | |
Controls | D33 | 14 | NA | C |
F38 | 16 | NA | C | |
A26 | 14 | 247 | C | |
9959 | 14 | 34, 74, 106, 133 | C | |
9 | 14 | 267 | C | |
B23 | 14 | NA | C |
NA, not applicable; DPC, days after challenge; C, challenge strain (dominant); V, vaccine strain (dominant); c, challenge strain; v, vaccine strain.
*Pony was killed at this date because of severe disease.