Gu and Roizman. 10.1073/pnas.0707266104. |

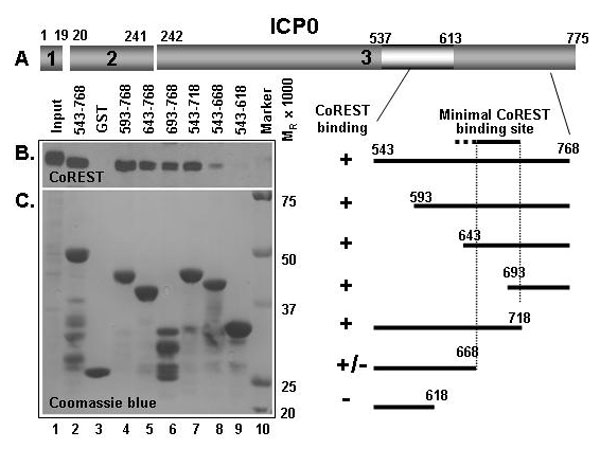
Fig. 7. Identification of ICP0 residues interacting with CoREST. To fine-map the binding site for CoREST, a series of deletions from both 5′ and 3′ sequences of the encoding ICP0543-768 protein were generated. The plasmids encoding the truncated proteins were fused in-frame with GST, expressed, and purified with glutathione Sepharose beads. GST-ICP0 pull-down assay was carried out as described in Experimental Procedures. The precipitates were electrophoretically separated on denaturing gels and reacted with anti-CoREST antibody. (B) Immunoblot with anti-CoREST antibody. (C) Coomassie blue-stained PVDF membrane. The dashed lines indicate the boundaries of the binding site.
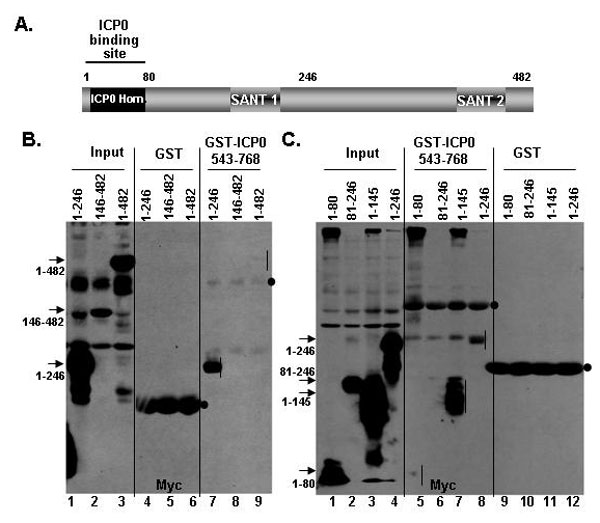
Fig. 8. CoREST 1-145 aa bind to ICP0. (A) Position of ICP0 homology site (residues 3-79) and key motifs in CoREST. (B and C) HEK 293 cells were transfected with plasmids encoding truncated polypeptides of CoREST tagged with Myc. After 40 h of incubation, the cells were harvested and lysed. The cell lysates were reacted with GST or GST-ICP0543-768 polypeptides bound to glutathione Sepharose beads as described above. The precipitates were solubilized, electrophoretically separated, and reacted with monoclonal anti-Myc antibody. Ectopically expressed CoREST polypeptides pulled down by the GST-ICP0543-768 polypeptide are indicated by vertical bars. GST and GST-ICP0543-768 reacted with anti-Myc antibody are marked by dots. The truncated CoREST polypeptides in input lanes are identified by arrows.
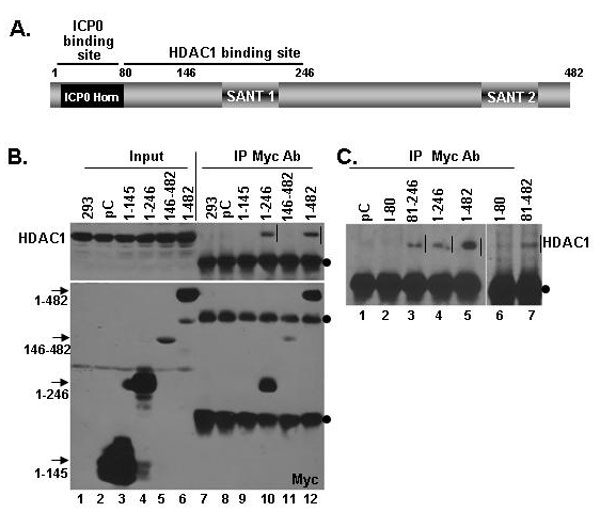
Fig. 9. The binding site for HDAC 1 on CoREST is between residues 81 and 246. Plasmids encoding truncated CoREST proteins described above were transfected into HEK 293 cells. After 40 h of incubation, the cells were harvested and lysed. The total cell lysates were reacted with anti-Myc antibody. The precipitates collected with protein A Sepharose beads were electrophoretically separated on denaturing gels and immunoblotted with anti-HDAC1 antibody. The results indicate that the HDAC1 binding site is located between residues 81 and 246. HDAC1 pulled down by CoREST deletions are indicated by vertical bars. The antibody heavy and light chains are marked by dots. The positions of the truncated CoREST polypeptides in input lanes are indicated by arrows.
Table 1. Primers and their sequences in deletion constructs of CoREST with N-terminal Myc tag
Plasmid | Deletion | Primer | |
pRB8502/pRB8512 | 1-482 | CoR7 | 5′-GCAAGGCCTATGGTGGAGAAGGGCCCCGAGGTCTCAGGGAAGCG-3′ |
CoR10 | 5′-GCACTGCAGTCAGGAGGCAGATGCATATCTGACATCCAGAACAGG-3′ | ||
pRB8503/pRB8513 | 1-80 | CoR7 | 5′-GCAAGGCCTATGGTGGAGAAGGGCCCCGAGGTCTCAGGGAAGCG-3′ |
CoR19 | 5′-GCACTGCAGTCAGCTGCTGTTGCCATTGGGCG-3′ | ||
pRB8504/pRB8514 | 81-246 | CoR20 | 5′-GCAAGGCCTAGCAACTCCTGGGAGGAAGG-3′ |
CoR12 | 5′-GCACTGCAGTCAGCATGGCGATCCATCACAC-3′ | ||
pRB8505/pRB8515 | 1-145 | CoR7 | 5′-GCAAGGCCTATGGTGGAGAAGGGCCCCGAGGTCTCAGGGAAGCG-3′ |
CoR11 | 5′-GCACTGCAGTCACTTTGCTTCTGACAGATTTTG-3′ | ||
pRB8506/pRB8516 | 1-246 | CoR7 | 5′-GCAAGGCCTATGGTGGAGAAGGGCCCCGAGGTCTCAGGGAAGCG-3′ |
CoR12 | 5′-GCACTGCAGTCAGCATGGCGATCCATCACAC-3′ | ||
pRB8507/pRB8517 | 146-482 | CoR3 | 5′-GCAAGGCCTTTGGATGAATACATTGC-3′ |
CoR10 | 5′-GCACTGCAGTCAGGAGGCAGATGCATATCTGACATCCAGAACAGG-3′ | ||
pRB8508/pRB8518 | 81-482 | CoR20 | 5′-GCAAGGCCTAGCAACTCCTGGGAGGAAGG-3′ |
CoR10 | 5′-GCACTGCAGTCAGGAGGCAGATGCATATCTGACATCCAGAACAGG-3′ | ||
pRB8509/pRB8519 | 247-482 | CoR18 | 5′-GCAAGGCCTGCCCGGAAACAAAAACG-3′ |
CoR10 | 5′-GCACTGCAGTCAGGAGGCAGATGCATATCTGACATCCAGAACAGG-3′ |
Table 2. Primers and their sequences in deletion constructs of ICP0
Plasmid | Deletion | Primer | Sequence |
pRB8401 | 543 -768 | ICP0 7 | 5 ′-CGAGAATTCCGCGGCCAGGGTGGGCCCG-3′ |
ICP0 11 | 5 ′-CGACTCGAGCGACGCCCCCTGCTCCCCGGAC-3′ | ||
pRB8402 | 593 -768 | ICP0 8 | 5 ′-CGAGAATTCGGGGCCGTCGGTGCCCTGGGAG-3′ |
ICP0 11 | 5 ′-CGACTCGAGCGACGCCCCCTGCTCCCCGGAC-3′ | ||
pRB8403 | 643 -768 | ICP0 9 | 5 ′-CGAGAATTCCTGCCCATCTCGGGGGTCTC-3′ |
ICP0 11 | 5 ′-CGACTCGAGCGACGCCCCCTGCTCCCCGGAC-3′ | ||
pRB8404 | 693 -768 | ICP0 10 | 5 ′-CGAGAATTCCGGCTGCGGGCCGCGGTCCCCG-3′ |
ICP0 11 | 5 ′-CGACTCGAGCGACGCCCCCTGCTCCCCGGAC-3′ | ||
pRB8405 | 543 -718 | ICP0 7 | 5 ′-CGAGAATTCCGCGGCCAGGGTGGGCCCG-3′ |
ICP0 12 | 5 ′-CGACTCGAGGGGGGGCATCACGTGGTTACC-3′ | ||
pRB8406 | 543 -668 | ICP0 7 | 5 ′-CGAGAATTCCGCGGCCAGGGTGGGCCCG-3′ |
ICP0 13 | 5 ′-CGACTCGAGGGGCAGGCAGTCCCCCGTGATAG-3′ | ||
pRB8407 | 543 -618 | ICP0 7 | 5 ′-CGAGAATTCCGCGGCCAGGGTGGGCCCG-3′ |
ICP0 14 | 5 ′-CGACTCGAGCGGCCCCCGCGGCCCAGAAGC-3′ |