[View Larger Version of this Image]
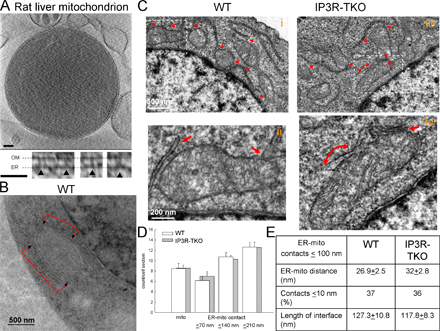
Figure S1. Close ER-mitochondria associations in isolated liver mitochondria and in wild-type and IP3R-TKO DT40 cells. (A) Tomographic reconstruction of a frozen-hydrated rat-liver mitochondrion (1,200-nm diameter) with large ER vesicle attached. (top) Slice (1.8 nm thick) from the tomogram, showing ER membrane wrapped around the upper right quadrant of the mitochondrion. The ER is attached at the top and, more extensively, at the lower region indicated by the dashed rectangular box. Bar, 100 nm. (bottom) Examples of tethers connecting ER and OM from various slices inside the subvolume defined by the dashed box. Bar, 50 nm. The membrane profiles and tethers do not appear as sharp as those in Fig. 1 A because the overall dimensions of this mitochondrion are much greater than those of the mitochondrion of Fig. 1 A, resulting in somewhat reduced resolution in the reconstruction. (B) Micrograph of a quick-frozen wild-type DT40 cell (WT) showing ER flanking a mitochondrion. (C) Micrographs of chemically fixed WT and IP3R-TKO showing numerous associations between ER and mitochondria. (D) Dimensions of the ER-mitochondria interface in chemically fixed wild type and IP3R-TKO. Mitochondria were counted and the smallest distance between mitochondria, and adjacent ER stacks were measured and sorted into 70-nm bins (11 cells for each). Similar results were obtained in the quick-frozen wild type, except that because of the weak staining, the measurements of the smooth ER-mitochondria distance was not feasible. (E) ER-mitochondria distance and interface area for the associations (~100 nm gap; 150 associations in each condition).