Blood, Vol. 111, Issue 2, 856-864, January 15, 2008
The multiple myeloma–associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity
Blood Lauring et al. 111: 856
Supplemental materials for: Lauring et al
Files in this Data Supplement:
- Table S1. Primer sequences for real-time PCR and gene targeting (PDF, 56.1 KB)
- Table S2. Relative expression levels of selected adhesion molecules in MMSET-knockdown KMS-11 cells (PDF, 47.7 KB) -
Quantitative real-time PCR was performed on cDNA prepared from empty vector lentivirus- or MMSET shRNA lentivirus-transduced KMS-11 cells. Expression for each gene was normalized to GAPDH expression levels. Control cell gene expression was set at 1 and the level in MMSET shRNA cells is expressed as a ratio relative to controls. - Figure S1. PCR-based screening for homologous recombination-mediated disruption of MMSET (JPG, 48 KB) -
(A) Schematic of gene targeting construct. 5′ and 3′ arms homologous to the introns on either side of MMSET exon 7 flank a drug selection cassette consisting of a promoter-less neomycin resistance gene (Neo) with a splice acceptor (SA), internal ribosome entry site (IRES), and flanking loxP sites (triangles). (B) Homologous targeting replaces exon 7 with the neomycin cassette, which can be screened for using primers 1 and 2 (right panel). A similar PCR screen was performed across the 3′ homology arm (not shown). (C) Expression of Cre recombinase deletes the neomycin cassette, leaving a single loxP site in place of the deleted exon 7. PCR using primers 1 and 3 detects intact MMSET (right panel, upper band) as well as a shorter product from the targeted, Cre’d allele (right panel, lane 2).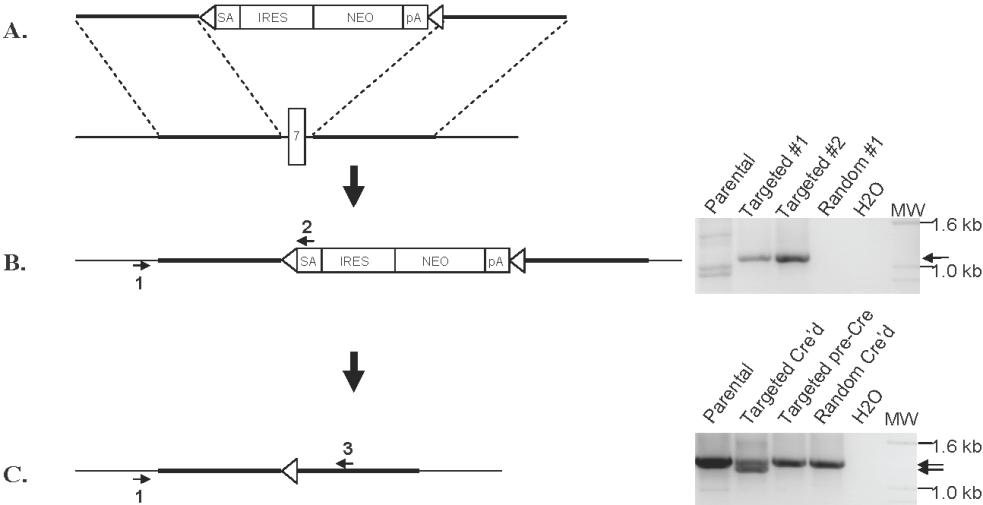
- Figure S2. RT-PCR for IgH-MMSET fusion transcripts discriminates clones with disruption of the translocated and non-translocated MMSET alleles (JPG, 34.4 KB) -
RT¬PCR was performed on the indicated KMS-11 clones after Cre-loxP excision of the neomycin targeting cassette using the indicated primers. A smaller product is detected in the T-KO cells, which have fusion transcripts lacking exon 7 (lane 2). Non-T-KO cells have fusion transcripts with an intact exon 7 (lanes 4 and 5), as do random integrant clones (lanes 1 and 3).
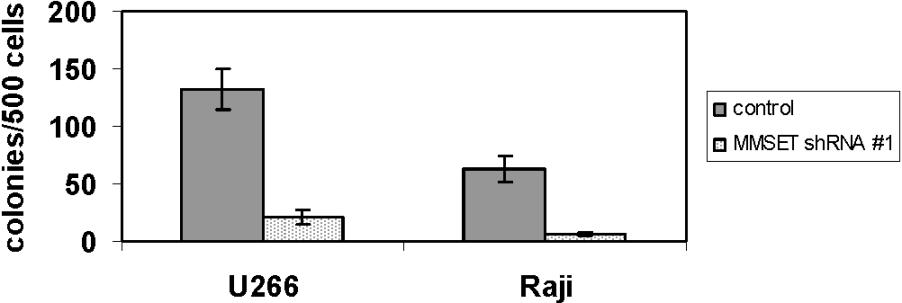
- Figure S3. MMSET knockdown reduces methylcellulose colony formation in non-t(4;14) U266 MM cells and Raji Burkitt lymphoma cells (JPG, 38.3 KB) -
Standard devations are shown.