| As splicing of most pre-mRNAs occurs co-transcriptionally, peripheral movements were strictly dependent on ongoing RNA polymerase II transcription. If inhibitors of RNA polymerase II were added, neither peripheral extensions nor (dis)associating particles were observed. |
 | The surrounding borderline of the detected speckle is coloured in green. The RNA polymerase II inhibitor caused a marked rounding up of the speckles and no peripheral movements were observed. QuickTime Video |
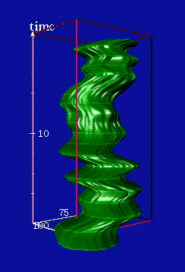 | Reconstructed 3D spatio-temporal structure of the speckle based on the extracted borderline of the 20 images (time lapse: 1 min). The highlighted rings along the reconstructed shapes correspond to outlines of the speckle at discrete time steps. QuickTime Video |
 | 3D time-space reconstruction from a speckle corrected for the speckle movement. QuickTime Video |
[ motility of secretory vesicles ]
[ simulation of vesicle dynamics and tracking ]
| | Get QuickTime 3 to view the movies |