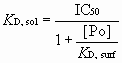 [1]
[1]Wang et al. 10.1073/pnas.0712326105. |
Fig. 8. MALDI-TOF mass spectrum for 15.
Fig. 9. MALDI-TOF mass spectrum for 17.
Fig. 10. Inhibition of 2G12 binding to normal-density Man4 slide at various concentrations of different glycodendrons.
Fig. 11. Inhibition of 2G12 binding to high-density Man4 slide at various concentrations of different glycodendrons.
Fig. 12. Flow-cytometric analysis of DC-SIGN expression. Jurkat cells were subsequently stained with anti-DC-SIGN antibody and fluorescent secondary antibodies. Red, mock-transfected Jurkat cells; green, pFLAG-CMV1-DC-SIGN-transfected Jurkat cells.
Table 4. Printed concentration for glycodendron 17 derivative and the corresponding dissociation constants on the surface
Printing concentration, mM | K D,surf, nM |
100 | 2.2 |
80 | 1.8 |
60 | 2.1 |
40 | 2.1 |
20 | 3.4 |
10 | 4.5 |
8 | 3.8 |
6 | 3.9 |
4 | 3.6 |
2 | 4.2 |
1 | 4.5 |
0.8 | 4.0 |
0.6 | 5.3 |
0.4 | 12 |
0.2 | 22 |
0.1 | 53 |
0.08 | 60 |
0.06 | 65 |
0.04 | 78 |
0.02 | 78 |
SI Materials and Methods
Syntheses of Compounds.
General. All chemicals were purchased from Aldrich or Acros and used without further purification. Reactions were monitored with analytical thin layer chromatography (TLC) in EM silica gel 60 F254 plates and visualized under UV (254 nm) and/or staining with acidic cerium ammonium molybdate or ninhydrin. Flash column chromatography was performed on silica gel 60 (35-75 mm, EM Science) or Iatrobeads 6RS-8060 (Mitsubishi Kagaku Iatron). 1H-NMR and 13C NMR spectra were recorded on a Bruker DRX-500 spectrometer at 20°C. 1H NMR spectra are reported in this order: chemical shift; multiplicity; coupling constant(s); number(s) of proton. The MALDI-TOF mass spectrometry was performed on a PerSeptive Biosystems Voyager-DE Biospectrometry workstation using 2,5-dihydroxybenzoic acid (DHB) as the matrix. The data were analyzed by Data Explorer software v.3.2.Boc-G1-alkyne, 7.
To a stirred solution of tri-acid compound 6 (0.47g, 0.93mmol), propargylamine (0.26g, 4.66mmol), HOBt (0.57g, 3.73mmol), and DIEA (0.81 ml, 4.66mmol) in DMF (10 ml) at 0°C was added EDC (0.89g, 4.66mmol). The reaction mixture was allowed to return to room temperature gradually and stirred overnight, which was followed by evaporating in vacuo and column chromatography (MeOH:CHCl3 = 1:20) to give purified product 7 (0.43g, 75%) as colorless oil.1
H NMR (500MHz, CDCl3) d 1.44 (s, 9H), 2.27 (t, J = 2.4Hz, 3H), 2.58-2.40 (m, 8H), 3.39 (dd, J1 = 12.1Hz, J2 = 5.9Hz, 2H), 3.69 (s, 6H), 3.73 (t, J = 5.7Hz, 6H), 4.06 (dd, J1 = 5.5Hz, J2 = 2.6Hz, 6H), 5.30 (s, 1H), 6.33 (s, 1H), 7.00 (s, 3H);13
C NMR (125MHz, CDCl3) d 28.28, 28.79, 36.15, 36.83, 37.03, 59.64, 67.08, 69.28, 71.19, 79.17, 79.95, 155.96, 171.28, 172.04;ESI-TOF HRMS calculated for C30H46N5O9 (M+H)+: 620.3290, found 620.3290.
NH2-G1-alkyne, 8.
To a flask containing compound 7 (0.14g, 0.22mmol) in CH2Cl2 (1 ml) was added TFA solution (1 ml, 50% vol/vol in CH2Cl2) at 0°C. After all starting material were consumed, the reaction mixture was evaporated in vacuo. The residual TFA in the mixture was removed by Amberlite IRA-743 ion-exchange resin. The crude product (0.11g, 96%) was used in the next step without further purification.1
H NMR (500MHz, CD3OD) d 2.43 (t, J = 5.9Hz, 6H), 2.58 (t, J = 2.4Hz, 3H), 2.62 (t, J = 6.4Hz, 2H), 3.16 (t, J = 6.1Hz, 2H), 3.60-3.75 (m, 12H), 3.96 (d, J = 2.2Hz, 6H);13
C NMR (125MHz, CD3OD) d 29.42, 33.32, 37.15, 61.56, 68.35, 69.87, 72.27, 80.75, 172.18, 173.61;ESI-TOF HRMS calculated for C25H38N5O7 (M+H)+: 520.2766, found 520.2779.
Boc-G2-alkyne, 9.
To a stirred solution of tri-acid compound 6 (0.022g, 0.043mmol), compound 8 (0.092g, 0.176mmol), HOBt (0.02g, 0.132mmol), and DIEA (0.03 ml, 0.176mmol) in DMF (1 ml) at 0°C was added EDC (0.042g, 0.22mmol). The reaction mixture was allowed to return to room temperature gradually and stirred 24 h, which was followed by evaporating in vacuo and column chromatography (MeOH:CHCl3 = 1:5) to give purified product 9 (0.084g, 97%) as colorless gum.1
H NMR (500MHz, CD3OD) d 1.51 (s, 9H), 2.44-2.62 (m, 32H), 2.70 (t, J = 2.6Hz, 9H), 3.36 (t, J = 6.6Hz, 2H), 3.50 (t, J = 7.0Hz, 6H), 3.72-3.87 (m, 48H), 4.06 (d, J = 2.2Hz, 18H);13
C NMR (125MHz, CD3OD) d 28.88, 29.49, 37.21, 37.32, 37.58, 37.86, 38.25, 47.88, 61.49, 61.58, 68.47, 68.71, 70.04, 72.44, 80.15, 80.90, 158.16, 173.59, 173.68, 173.85, 173.96;ESI-TOF HRMS calculated for C96H142N17O30 (M+H)+: 2013.0103, found 2013.0113.
NH2-G2-alkyne, 10.
The same procedure as for compound 8, (99%).1
H NMR (500MHz, CD3OD) d 2.50-2.35 (m, 32H), 2.62 (t, J = 2.4Hz, 9H), 3.15-3.24 (m, 2H), 3.41 (t, J = 7.0Hz, 6H), 3.63-3.78 (m, 48H), 3.98 (d, J = 2.6Hz, 18H);13
C NMR (125MHz, CD3OD) d 29.47, 37.17, 37.26, 37.44, 61.47, 61.58, 68.44, 68.64, 69.99, 72.44, 80.89, 172.33, 173.57, 173.64, 173.84;ESI-TOF HRMS calculated for C91H134N17O28 (M+H)+: 1912.9584, found 1912.9564.
Boc-G3-alkyne, 11.
To a stirred solution of tri-acid compound 6 (5.9 mg, 0.012mmol), compound 10 (0.079g, 0.041mmol), HOBt (8.0 mg, 0.051mmol), and DIEA (0.013 ml, 0.072mmol) in DMF (0.8 ml) at 0°C was added EDC (0.014g, 0.072mmol). The reaction mixture was allowed to return to room temperature gradually and stirred 72 h. which was evaporated in vacuo. The residue was purified by silica gel column chromatography (MeOH:CHCl3 = 1:2) and followed by another size-exclusion column (Bio-gel P-10, Bio-Rad) to give purified product 11 (0.0285g, 40%) as colorless gum.1
H NMR (500MHz, CD3OD) d 1.52 (s, 9H), 2.44-2.56 (m, 104H), 2.72 (t, J = 2.6Hz, 27H), 3.50 (t, J = 7.0Hz, 26H), 3.72-3.85 (m, 156H), 4.07 (d, J = 2.6Hz, 54H);13
C NMR (125MHz, CD3OD) d 29.01, 29.54, 37.25, 37.35, 37.62, 61.52, 61.54, 68.52, 68.76, 70.08, 72.53, 80.99, 81.01, 173.60, 173.63, 173.67, 173.80;MALDI-TOF calculated for C294H430N53O93 (M+H)+: 6191, found 6192.
Representative procedures for conjugating oligomannose to alkynyl dendron via CuAAC reaction (Boc-Gn-Alkyne ®Boc-Gn-Manx), 13~18
. To a stirred solution of Man9-N3 (5.0 mg, 3.14mmol) and Boc-G3-Alkyne (0.57 mg, 0.09mmol) in 0.2 ml H2O was added aqueous CuSO4 (20 mM, 10 ml), triazole ligand 12 (20 mM in DMSO, 10 ml) and sodium ascorbate (20 mM, 20 ml). The mixture was stirred for 2 h, and analyzed by MALDI-TOF MS to confirm the completeness of the reaction. The mixture was then repeatedly centrifugal filtered (Millipore Microcon YM-3) and washed to give white solid as product 18 (4.3 mg, 98%) after concentrated.Boc-G1-Man4, 13.
1H NMR (500MHz, D2O) d 1.13-1.27 (m, 15H), 1.41-1.52 (m, 6H), 1.72-1.80 (m, 6H), 2.18 (t, J = 5.9Hz, 2H), 2.35 (t, J = 5.7Hz, 6H), 3.10 (t, J = 6.3Hz, 2H), 3.33-3.92 (m, 90H), 4.26 (t, J = 7.0Hz, 6H), 4.32 (s, 6H), 4.66 (s, 3H), 4.91 (s, 3H), 5.16 (s, 3H), 5.21 (s, 3H), 7.77 (s, 3H).MALDI-TOF calculated for C117H198N14O72Na (M+Na)+: 2974, found 2975.
Boc-G2-Man4, 14.
1H NMR (500MHz, D2O) d 1.14-1.28 (m, 27H), 1.46-1.54 (m, 18H), 1.66-1.82 (m, 18H), 2.27-2.32 (m, 8H), 2.34-2.41 (m, 24H), 3.13-3.17 (m, 2H), 3.25-3.30 (m, 6H), 3.37-4.02 (m, 282H), 4.28 (t, J = 6.8Hz, 18H), 4.35 (s, 18H), 4.70 (s, 9H), 4.95 (s, 9H), 5.20 (s, 9H), 5.25 (s, 9H), 7.81 (s, 9H).MALDI-TOF calculated for C357H600N44O219Na (M+Na)+: 9031, found 9028.
Boc-G3-Man4, 15.
MALDI-TOF calculated for full conjugation C1077H1807N134O660 (M+H)+: 27178. Also see SI Fig. 8 for the full spectrum.Boc-G1-Man9, 16.
1H NMR (500MHz, D2O) d 1.15-1.25 (m, 15H), 1.44-1.52 (m, 6H), 1.73-1.80 (m, 6H), 2.18 (t, J = 5.9Hz, 2H), 2.36 (t, J = 5.3Hz, 6H), 3.10 (t, J = 5.9Hz, 2H), 3.32-3.99 (m, 180H), 4.26 (t, J = 6.8Hz, 6H), 4.32 (s, 6H), 4.71 (s, 3H), 4.91 (s, 9H), 5.01 (s, 3H), 5.17 (s, 3H), 5.20 (s, 3H), 5.26 (s, 3H), 7.77 (s, 3H).MALDI-TOF calculated for C207H348N14O147Na (M+Na)+: 5405, found 5405.
Boc-G2-Man9, 17.
MALDI-TOF calculated for full conjugation C627H1051N44O444 (M+H)+: 16301, found 16298. Also see SI Fig. 9 for the full spectrum.Boc-G3-Man9, 18.
MALDI-TOF calculated for full conjugation C1887H3157N134O1335 (M+H)+: 49055. Also see Fig. 4 for the full spectrum.Microarray Experiments.
Microarray Fabrication. Man4 on normal-NHS-density slide. NHS-coated glass slides (slide H, Schott North American) were printed by robotic pin deposition of ~0.7 nl of Man4, 1, with concentrations of 100, 75, 50, 40, 30, 20, 15, 10, 7.5, 5, 3, 1, 0.75, 0.5, 0.25, and 0.1 mM in print buffer [300 mM phosphate (pH 8.5), containing 0.005% Tween 20] from left to right with 16 replicates vertically placed in each subarray and there were totally 16 replicates of subarrays on one slide. After the solvent of the printed matrix evaporated, the slide was washed with PBST (0.05% Tween 20) buffer and then treated with blocking solution (superblock blocking buffer in PBS; Pierce) at room temperature for 1 h. The slides were then washed with PBS buffer, dried, and stored in dessicator.Man4 on high-NHS-density slide.
Similar procedure as for normal density slides was used. The NHS activated slides were manufactured by GE Healthcare (CodeLink HD). The printing concentration for Man4, 1, was 100, 80, and 60 mM.Second-generation Man9 dendron 17 on normal-NHS-density slide.
Similar procedure as for normal density Man4 slide was used. Derivative of 17 with a linker connecting free amine was used for printing. The printing concentrations are 100, 80, 60, 40, 20, 10, 8, 6, 4, 2, 1, 0.8, 0.6, 0.4, 0.2, 0.1, 0.08, 0.06, 0.04, and 0.02 mM. The KD,surf of 2G12 complex was obtained (SI Table 4) as described in ref. 1. The limit of detection is defined as the lowest 2G12 concentration applied to slide that resulted in signal to noise ratio greater than 10.Determination of KD,surf of Glycodendron 17.
Similar procedure as described in ref. 1.Microarray Competitor Assay for 2G12 Complex: KD,sol determination.
The solution equilibrium dissociation constant (KD,sol) for oligomannose-2G12 complex interactions can be determined using microarrays in a competitive assay (1). The equation that describes the binding of the two ligands to the same site on the protein is identical to that for the competitive inhibition of an enzyme-catalyzed reaction (2). It is possible to take advantage of the convenience of IC50 measurements and still report inhibitory potency in terms of true KD,sol values. The final forms of the relationship can be simply presented as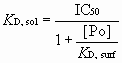 [1]
[1]
The limit of detection is defined as the lowest 2G12 concentration applied to slide that resulted in signal to noise ratio greater than 10. For the 17 immobilized slide, the limit was found at 0.05 mg/ml, where for the normal Man4 slide is 3 mg/ml.
IC50 Determined by the Microarray Competitor Assay for Fc-DC-SIGN.
1.5 ml of serial diluted of competitors were mixed with 1.5 ml of 80 mg/ml Fc-DC-SIGN, under buffered condition (binding buffer: 2 mM CaCl2, 2 mM MgCl2, 150 mM NaCl, 0.05% Tween 20, 1% BSA, 20 mM Tris-HCl, pH 7.4). The mixtures (3 ml) were applied directly to each subarray. After incubation in a humidified chamber for 1 h at room temperature, it was rinsed sequentially with PBS, PBS-T buffer (0.05% Tween 20 in PBS), and distilled water. After residual water was removed, 15 ml of Cy3-labeled goat anti-human IgG antibody (0.01 mg/ml in binding buffer) was applied to each submatrix and incubated in moisture chamber for 30 min. The following washing and analyzing process was as described for 2G12 complex experiments.Competition ELISA for Fc-DC-SIGN.
DC-SIGN competitions were done with 1.5 mg/ml Fc-DC-SIGN as described above with a few modifications because DC-SIGN is a C-type lectin that requires Ca2+ for binding. A Tris (10 mM, pH 7.8) buffer containing NaCl (150 mM), i.e., TBS, replaced PBS in the above-mentioned buffers used for coating, blocking, and washing. After the initial wash step, CaCl2 (10 mM) was included in the blocking solution and all subsequent incubation and wash buffers until substrate addition. DC-SIGN-Fc binding was detected with an alkaline phosphatase-conjugated goat anti-human IgG, Fcg-specific, Ab (Jackson ImmunoResearch Laboratories) diluted 1:1,000.Overexpression of Cell-Surface DC-SIGN.
Full-length cDNA encoding human DC-SIGN was PCR-amplified and subcloned into pFLAG-CMV1. The construct pFLAG-CMV1-DC-SIGN was transfected to Jurkat cells by a MicroPorator (NanoEnTek) according to the instructions. Briefly, Jurkat cells (2 ´ 106) were mixed with 10 mg of plasmid in 100 ml of resuspension buffer and electroporated once with a pulse voltage of 1,410 V and a pulse width of 30 ms. Cells were cultured in complete growth medium with no antibiotics for 2 days before surface staining.Culture of Monocyte-Derived Dendritic Cells (MDDCs).
Peripheral blood mononuclear cells (PBMCs) were isolated from white blood cell concentrates (obtained from San Diego Blood Bank, CA) by standard density gradient centrifugation with Ficoll-Paque (GE Healthcare). Monocytes were then purified with anti-CD14 microbeads (Miltenyi Biotec) and cultivated in RPMI medium 1640 (Invitrogen) supplemented with 10% FBS (Invitrogen), 800 units/ml human GM-CSF (PeproTech), and 500 units/ml human IL-4 (PeproTech) for 6 days to differentiate to MDDCs.Flow-Cytometric Analysis.
Cells were stained with fluorescein-conjugated glycodendrons (50 pmol for 3 ´ 105 cells) or mouse anti-DC-SIGN mAb (clone 120507; R&D Systems) in FACS buffer (1% FBS and 0.1% NaN3 in PBS) at 4°C for 20 min. Phycoerythrin (PE)-conjugated goat anti-mouse IgG was used to stain DC-SIGN subsequently at 4°C for 20 min. Fluorescence intensity was analyzed by a LSR II (BD Biosciences) and CellQuest Pro (BD Biosciences).1. Liang PH, Wang SK, Wong CH (2007) Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: Determination of surface and solution dissociation constants. J Am Chem Soc 129:11177-11184.
2. Copeland RA (1996) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis (Wiley, New York).