Supplementary material for Adams et al. (1999) Proc. Natl. Acad. Sci. USA 96 (24), 13863-13868.
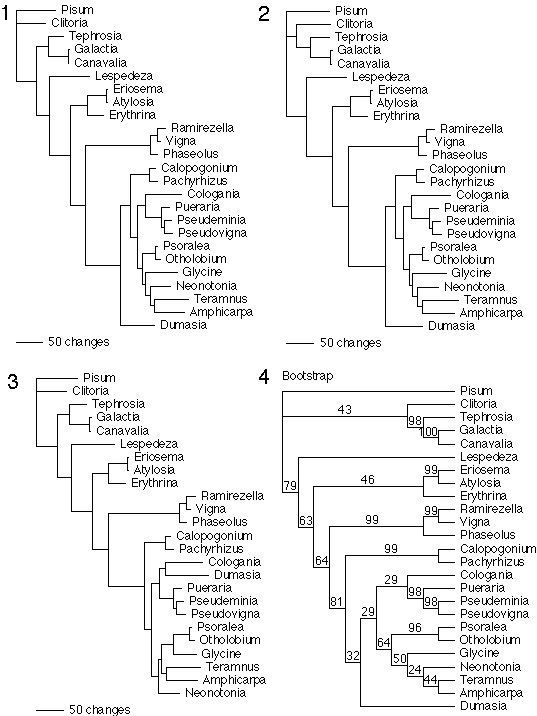
Fig. 6.
Separate analyses of rbcL and restriction fragment length polymorphism data (see below) provided low resolution and low support, especially within the Glycininae and allies. Thus, the rbcL and restriction fragment length polymorphism data were combined, in addition to adding partial ndhF sequences from some taxa. The results of analyses of individual data sets were congruent with the combined analyses (described below). Simultaneous parsimony analysis of four chloroplast genome data sets: (i) full rbcL gene sequences (1484 aligned nucleotide positions); (ii) partial ndhF gene sequences (670 aligned nucleotide positions, at the 3' end of ndhF); (iii) restriction site maps of the inverted repeat (IR) region (202 sites); and (iv) restriction site maps of the large single copy (LSC) region (355 sites). Not all data were available for all taxa analyzed. Data analyzed for each taxon: rbcL, ndhF, IR, and LSC: Vigna, Pachyrhizus, Cologania, Pseudeminia, Psoralea, Otholobium, Glycine, Neonotonia, Amphicarpa, Dumasia; rbcL, IR, and LSC: Erythrina, Calopogonium, Pueraria, Pseudovigna; IR and LSC: Ramirezella, Phaseolus; rbcL and IR: Clitoria, Tephrosia, Canavalia, Lespedeza, Atylosia; IR only: Galactia, Eriosema; rbcL only: Pisum. Heuristic analysis using 1,000 random taxon entry sequences and TBR branch swapping options in PAUP beta-1 identified three equally most parsimonious trees (trees 1-3) of length 1,426, with a consistency index of 0.68 (0.53 excluding uninformative characters) and a retention index of 0.57. The three trees differ in the placement of Dumasia and in the relationships among Glycine, Neonotonia, and Otholobium/Psoralea. However, the trees agree in most details with respect to hypothesized inactivations of cox2 genes. Tree 1 is shown in Fig. 5 of the text. Bootstrap analysis (shown in tree 4) was performed using by 1,000 replicates, 10 random taxon entry sequences, and TBR branch swapping in PAUP beta-1.