Supplementary material for Greally et al. (1999) Proc. Natl. Acad. Sci. USA 96 (25), 14430-14435.
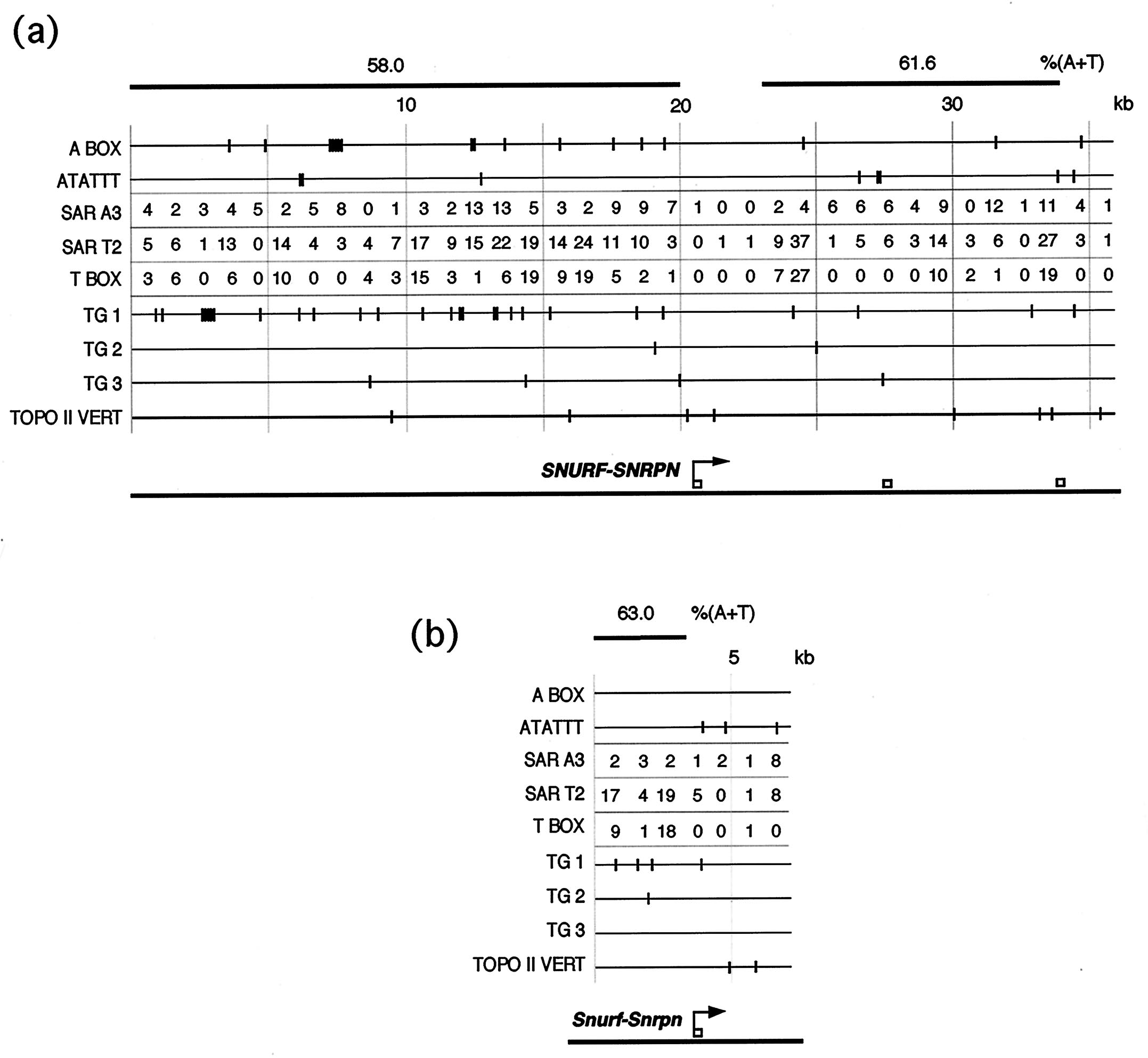
Fig. 9.
(a) The motifs found to characterize MARs [Greally, J. M., Guinness, M. E., McGrath, J. & Zemel, S. (1997) Mamm. Genome8, 805-810] were plotted for the sequences available at the human SNURF-SNRPN locus. An elevated frequency of motifs occurs in the regions found to bind specifically in the NMBA: the 20 kb upstream from SNURF-SNRPN has numerous motifs, particularly the first 10 kb upstream, whereas the region at and immediately downstream from exon 1 has very few motifs, corresponding to the restriction fragments from 8-1-1 and 12-E-E found to bind less strongly in the NMBA. (b) A similar pattern is seen for the mouse Snurf-Snrpn locus. The frequency of motifs is even higher in this region than that we observed for the imprinted Igf2/H19 region [Greally, J. M., Guinness, M. E., McGrath, J. & Zemel, S. (1997) Mamm. Genome 8, 805-810]. The presence of MAR motifs in both species complements the results of the NMBAs.