Blood, Vol. 113, Issue 2, 338-346, January 8, 2009
Functional assessment of perforin C2 domain mutations illustrates the critical role for calcium-dependent lipid binding in perforin cytotoxic function
Blood Urrea Moreno et al. 113: 338
Supplemental materials for: Urrea Moreno et al
Perforin sequencing
The modifications with respect to the previously published method13 are the following:
For amplification of exons 2 and 3 of PRF1 gene, PCR conditions were 4 minutes at 94°C followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 59°C, 1 minute at 72°C, and then 10 minutes at 72°C. Primers for sequencing of exon 3 were those used for PCR plus these four: 5´-GTCACCACCCAGGACTGCTG-3´, 5´-ATGACCTCCTTGGCACCTGT-3´, 5´-GTGGATGCCTATGTTGACCT-3´, and 5´-GGGTTCCAGGGTGTAGTCCA-3´. Amplification of DNA (Advantage-GC PCR kit, Clontech,Mountain View, CA) was performed in a 50 µl assay with 5× PCR buffer, 5 µl GC Melt, 0.2 mM dNTP, 0.2 µM of each primer, and 1 µl of Taq polymerase (Amplitak DNA polymerase, Branchburg, NJ, USA). Cycle sequencing was done using the BigDye terminator kit (version 2; Applied Biosystems, Foster City, CA, USA). The DNA fragments were separated on an ABI Prism 3100 DNA Analyzer (Applied Biosystems). Sequences were compared with one published PRF1 sequence (GenBank accession no. M31951.1) using Chromas v2.3 software (Technelysium, Tewantin, Australia).
Files in this Data Supplement:
- Table S1. C2 structures identified by BLAST and modeled in this study (PDF, 20.8 KB)
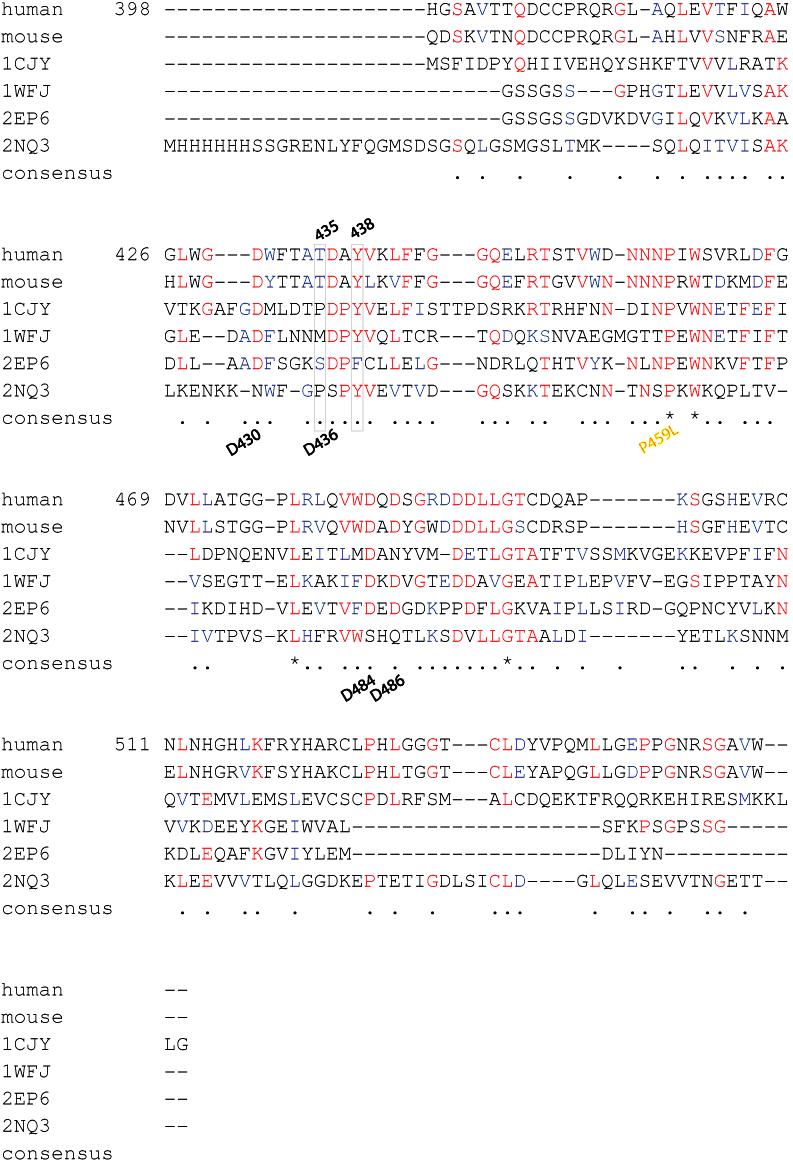
- Figure S1. Alignment of perforin C2 domains across species (JPG, 191 KB) -
Using t-coffee we aligned all available mammalian species to human perforin 398-555 C2 domain. Perfectly conserved residues are marked in the consensus sequence by (*); conserved residues are marked in the consensus by (.). The font color in each sequence illustrates conservation as well: highly conserved residues are in red font, moderately conserved residues in blue font, and non-conserved residues are in black font. Sheep was excluded from the alignment because of an extremely short C2 domain, ending two residues past Y438; however the sequence between T435 and Y438 was perfectly conserved with human perforin: TDAY. Gray box surrounds the T435 and Y438 discussed in manuscript.
- Figure S2. Alignment of type I C2 domains identified by sequence homology by BLAST (JPG, 211 KB) -
Using t-coffee we aligned to human perforin 398-555 C2 domain. Fonts and symbols are the same as described in Fig. S1. Gray box surrounds the T435 and Y438 discussed in manuscript. There are 12 perfectly conserved residues marked by an (*); 4 of these conserved residues are aspartates predicted to bind calcium (marked below the (*); 2 have been associated with FHLH2 when mutated (marked by yellow font below the (*)) (Ref 10).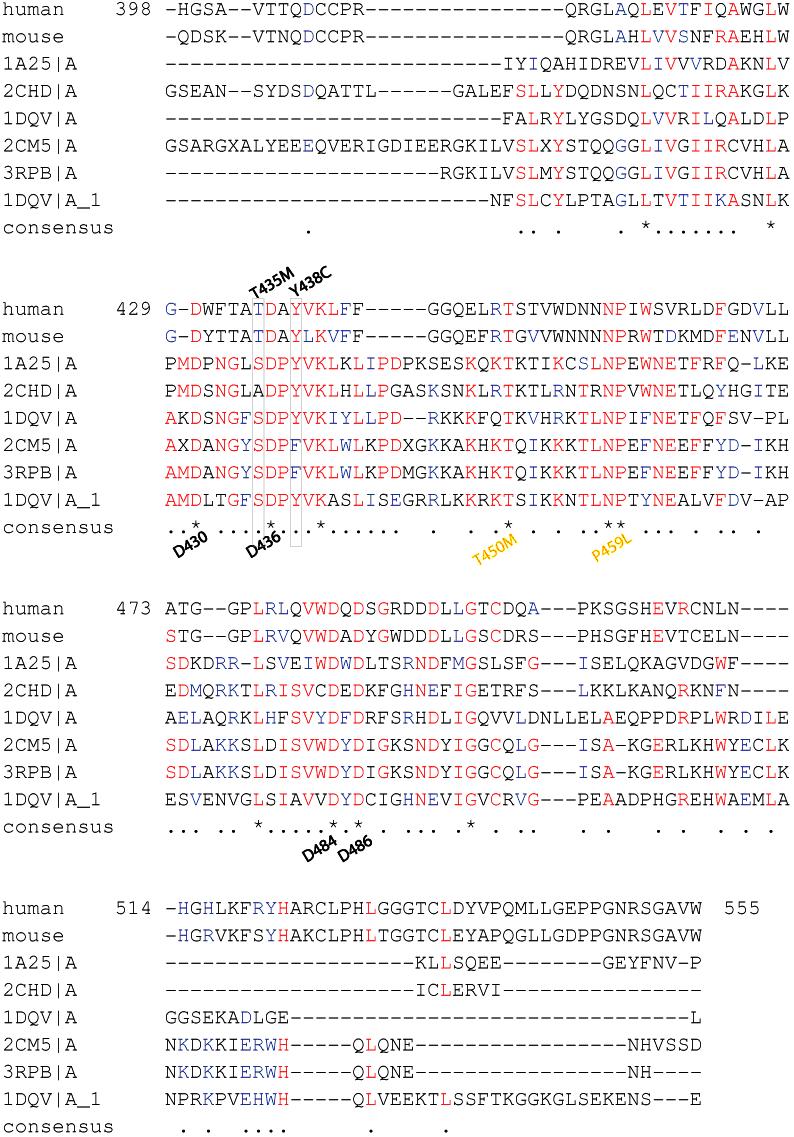
- Figure S3. Alignment of type II C2 domains identified by sequence homology by BLAST (JPG, 165 KB) -
Using t-coffee we aligned to human perforin 398-555 C2 domain. Fonts and symbols are the same as described in Fig. S1. Gray box surrounds the T435 and Y438 discussed in manuscript. There are 4 perfectly conserved residues marked by an (*); one of these residues has been associated with FHLH2 when mutated, yellow. The aspartates predicted to bind calcium are marked below the consensus (Ref 10).