Blood, Vol. 113, Issue 15, 3558-3567, April 9, 2009
Structure of the AML1-ETO eTAFH domain–HEB peptide complex and its contribution to AML1-ETO activity
Blood Park et al. 113: 3558
Supplemental materials for: Park et al
Comparison of eTAFH-HEB complex structure with docking models
The orientation of the HEB AD1 domain on eTAFH in our structure eminently disagrees with the docking models reported by two groups.1,2 In the previous studies, HEB peptide from G12 to F27 was assumed to be a single amphipathic α-helix, and the docking models were generated by HADDOCK program with identification of binding surface on eTAFH based on NMR footprint analysis and functional mutagenesis. Although the same strategy and method were used by both groups, the models showed a difference each other. In both docking models, the HEB peptide goes almost parallel to the axis of the binding groove formed by helix 1 and the loop between helix 1 and helix 4 of eTAFH while they have almost 180° orientation difference. On the other hand, a shorter fragment of HEB (D14-D22) is making contact with the binding groove of eTAFH, and the axis of the relatively short a-helix resides almost perpendicular to the same axis in our structure. Subsequent binding assay with shorter HEB peptide containing only T13-D22 revealed that this short fragment is sufficient to interact with eTAFH (Fig. 4A). A four-turn α-helix of HEB needed a longer binding groove parallel to the helix 1 and the loop between helix 3 and 4 of eTAFH in the docking experiments while the two and a half-turn α-helix is enough to fit the binding groove in our structure. The possible reasons of the discrepancy between our structure and previous docking models can be explained by the incorporation of T321 and F323 of eTAFH and S24 and F27 of HEB peptide as active residues in HADDOCK modeling1,2.
Files in this Data Supplement:
- Figure S1. Comparison of eTAFH:HEB complex with docking models (JPG, 48.6 KB) -
Orientation of the α-helix of HEB peptide are presented by cartoons (blue) on the experimental complex structure we have determined (A) and on the HADDOK models generated with PDB entry 2H7B1 (B) and 2PP42 (C). The differences are discussed in the main text.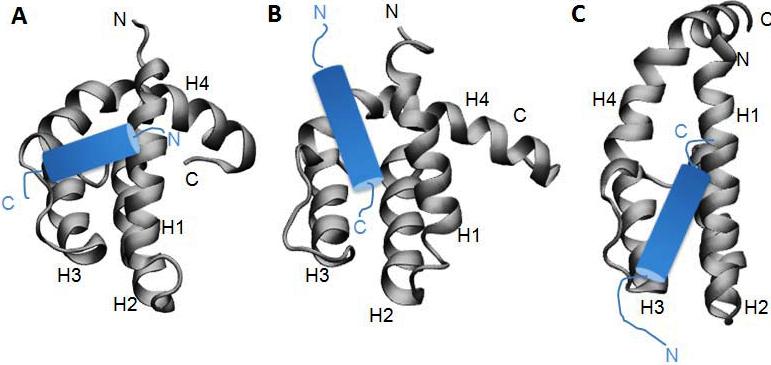
REFERENCES
1. Plevin, M.J., Zhang, J., Guo, C., Roeder, R.G. & Ikura, M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc. Natl. Acad. Sci. U S A 103, 10242-10247 (2006).
2. Wei, Y. et al. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat. Struct. Mol. Biol. 14, 653-661 (2007).