Detailed secondary structure models of LSU RNA from several species
These figures illustrate the structure of the LSU rRNA molecule for all major taxa. A conserved core can be found in all species. Only in some mitochondria with a higly reduced sequence (as shown in Fig. 5) helices from this core are missing. The core is interspersed with regions of extreme variability. These are indicated by red boxes and labeled V1 to V12. In eukaryotes, and especially in vertebrates, some of these regions are so variable, both in base composition and length, that no structure could be proposed based on comparative evidence.The stem helix which joins the 5' and 3' end of the molecule in Bacteria, plastids and most Archaea is labeled A1. Other structures branching from the central loop are labeled B to I. Helices are numbered in the 5' to 3' direction. Helices get a different number when they are separated by a multi branched loop in some species. The name of helices not belonging to the core structure but specific to certain taxa is constructed by adding an underscore and a number to the name of the preceding core helix.

Fig. 2. LSU rRNA secondary structure model for the bacterium Escherichia coli (Click on the picture for a high resolution view.)

Fig. 3. LSU rRNA secondary structure model for the archaebacterium Sulfolobus acidocaldarius (Click on the picture for a high resolution view.)
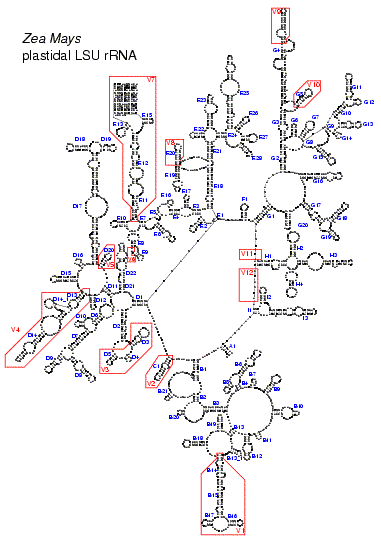
Fig. 4. LSU rRNA secondary structure model for the plastid of Zea mays (Click on the picture for a high resolution view.)

Fig. 5. LSU rRNA secondary structure model for the mitochondrion of Mus musculus (Click on the picture for a high resolution view.)
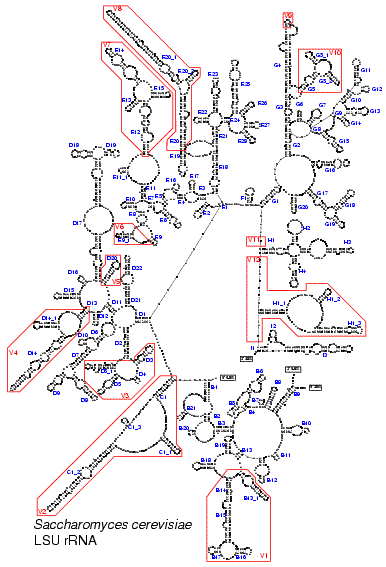
Fig. 6. LSU rRNA secondary structure model for the unicellular eukaryote Saccharomyces cerevisiae (Click on the picture for a high resolution view.)
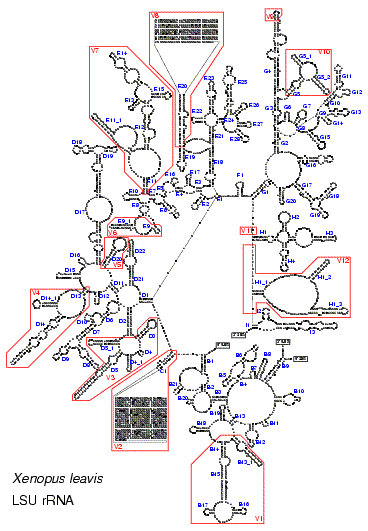
Fig. 7. LSU rRNA secondary structure model for the multicellular eukaryote Xenopus laevis (Click on the picture for a high resolution view.)