Supplementary material for Arora et al. (2001) Proc. Natl. Acad. Sci. USA 98 (13), 7241-7246. (10.1073/pnas.131132198)
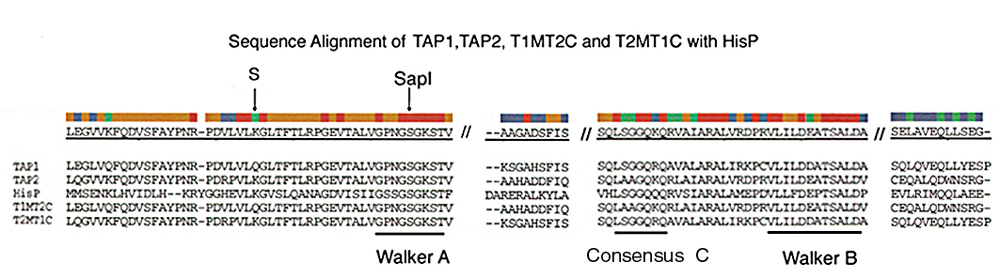
Fig. 6.
Sequence alignments with His P of the predicted NBD of TAP1, TAP2, T1MT2C, and T2MT1C. The predicted NBD sequences of TAP1, TAP2, T1MT2C, and T2MT1C are indicated and aligned with the HisP sequence. Locations of Walker A, Walker B, and consensus C sequence motifs are indicated. The position corresponding to the SapI restriction site that was used to generate T1MT2C and T2MT1C are indicated. S indicates the effective switch site in the TAP1 sequence relative to T1MT2C and in the TAP2 sequence relative to T2MT1C. To the left of S, T1MT2C is identical to the TAP1 sequence, and to the right of S, T1MT2C is identical to the TAP2 sequence. To the left of S, T2MT1C is identical to the TAP2 sequence, and to the right of S, T2MT1C is identical to the TAP1 sequence. The only regions shown C terminal to the Walker A motif are the clusters with highest sequence conservation and greatest sequence disparity between TAP1 and TAP2.