|
Caicedo and Schaal. 10.1073/pnas.0407899101. |
Fig. 3. Localities of the Solanum pimpinellifolium populations sampled for the study.
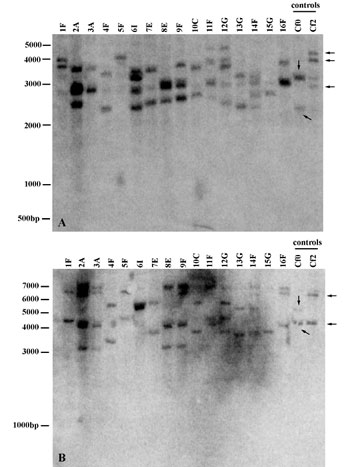
Fig. 4. Southern blots and hybridization results. Southern blots were prepared as described in ref. 1. Nylon membranes were incubated with the probe at 65°C overnight in high-SDS hybridization buffer (2). After hybridization, the membranes were washed twice for 10 min in 0.2´ SSC/0.1% SDS, also at 65°C. The probe used and the high stringency conditions of hybridization and washes were chosen to prevent binding to the related complex S. pimpinellifolium locus, Cf-9. Lanes correspond to individual plants from different populations. Individual names are collector numbers and do not refer to population of origin. The Solanum lycopersicum near-isogenic lines Cf2 (which contains introgressed Cf-2 and Hcr2-2A loci) and Cf0 (which contains Cf-2 homolog, Hcr2-0B, and Hcr2-2A homolog, Hcr2-0A) were used as controls (see ref. 3). Arrows in control lanes point to products corresponding to Cf-2.1, Cf-2.2, Hcr2-2A for Cf2, and Hcr2-0A and Hcr2-0B for Cf0 (see ref. 3). Of the 1,594-bp probe, 1,340 bp fall within the Cf-2.1 and Cf-2.2 coding regions. Neither restriction enzyme used cuts the probe. (A) DraI digest. DraI is predicted to cut Cf-2.1 in two sites flanking the probe, leading to a predicted band size of 4,617 bp. The enzyme is predicted to cut the following genes and their known flanking areas only in the region 5' of the probe: C.-2.2 (nine times): Hcr2-2A (three times), Hcr2-0A (five times), and Hcr2-0B (one time). (B) BclI digest. BclI is predicted to cut Cf-2.1 at two sites flanking the probe, leading to a predicted band size of 4,195 bp. The enzyme is predicted to cut Cf-2.2 and its known flanking region at two sites 5' of probe. BclI is not predicted to cut Hcr2-2A, Hcr2-0A, or Hcr2-0B.
1. Jeddeloh, J. A., Bender, J. & Richards, E. J. (1998) Gene Dev. 12, 1714-1725.
2. Church, G. M. & Gilbert, W. (1984) Proc. Natl. Acad. Sci. USA 81, 1991-1995.
3. Dixon, M. S., Hatzixanthis, K., Jones, D. A., Harrison, K. & Jones, J. D. G. (1998) Plant Cell 10, 1915-1925.
Table 4. Primers used for amplification and sequencing the Cf-2 homologs
|
Name |
Sequence |
Direction* |
|
Cf2p11 |
GATATGGTCCACTACTGTAGAGATG† |
f |
|
Cf2p14 |
CGTCTTCTTATTCTTGGCTCCTG† |
r |
|
Cf2p7 |
GAAAGCTATTACGATGACTCGGTGG |
f |
|
Cf2p7r |
CCACCGAGTCATCGTAATAGCTTTC |
r |
|
Cf2p9 |
GATGGATTACGTGGATATCCAG |
f |
|
Cf2p10 |
CTCGGCATATACAACACTTCCAGTG |
r |
|
Cf2p12 |
TCTCGGCATATACAACACTTCC |
r |
|
Cf2p13 |
TGCAATTTGACATCACTGGAAG |
f |
|
Cf2p15 |
GGAACTTTGCCAGAGCTGAGAG |
f |
|
Cf2paf |
CAACTTTGACTCAGTCCTTGG |
f |
|
Cf2par |
GATGACATCGACAAAACCTGA |
r |
|
Cf2p19 |
GTTGCATTTGCTTCGACTGA |
f |
|
Cf2p21 |
AGCCAAGCTTCAGATCATCC |
f |
|
Cf2p23 |
TGGCAATATGAGCAACTTGG |
f |
|
Cf2p9r |
CTGGATATCCACGTAATCCATC |
r |
|
Cf2p15r |
CTCTCAGCTCTGGCAAAGTT |
r |
|
Cf2p23r |
CCAAGTTGCTCATATTGCCA |
r |
|
Cf2p1 |
CTCTTACTTATCTAGATTTGAGTAATAACTCC |
f |
|
Cf2p21r |
GGATGATCTGAACCTTGGAC |
r |
*f, forward; r, reverse.
†
Primers used for amplification.Table 5. Pairwise similarity levels of A and B type leucine-rich repeat (LRR) units within Cf-2 homolog size classes
|
A type LRRs |
B type LRRs |
|||||
|
Homolog size class |
No. of repeats* |
Percent base pair similarity† |
Percent amino acid similarity‡ |
No. of repeats |
Percent base pair similarity |
Percent amino acid similarity |
|
Cf-2 |
9 |
75-100 |
58.3-100 |
11 |
75-100 |
54.2-100 |
|
Hcr2-p1 |
8 |
76.4-97.2 |
66.7-100 |
7 |
87.5-97.2 |
66.7-100 |
|
Hcr2-p2 |
5 |
80.6-98.6 |
70.8-95.8 |
4 |
83.3-98.6 |
70.8-95.8 |
|
Hcr2-p3 |
5 |
83.3-98.6 |
70.8-95.8 |
4 |
84.7-97.2 |
75-95.8 |
|
Hcr2-p4 |
4 |
83.3-98.6 |
70.8-95.8 |
3 |
83.3-100 |
70.8-100 |
|
Hcr2-p5 |
3 |
83.3-90.3 |
75-83.3 |
2 |
81.9 |
66.7 |
|
Hcr2-p6 |
2 |
81.9 |
75 |
1 |
NA |
NA |
|
Hcr2-p7 |
6 |
75-93.1 |
58.3-95.8 |
7 |
75-97.2 |
54.2-95.8 |
|
Hcr2-p8 |
2 |
97.2 |
91 |
2 |
90.2 |
79 |
NA, not applicable.
*Number of LRR repeats within the given Cf-2 homolog size class.
†
Percent nucleotide similarity between LRR units within a homolog size class.‡
Percent amino acid similarity between LRR units within a homolog size class.Fig. 5. Gene trees obtained with parsimony analysis in PAUP*, Ver. 4.0b3 (Sinauer, Sunderland, MA), for various partitions of the dataset. Heuristic searches used tree-bisection-reconnection (TBR), Collapse, and Multitrees options, with steepest descent not in effect. The root of the trees is unknown, and phylograms are shown here with midpoint rooting. For datasets in which more than one tree was found, a strict consensus cladogram is shown. (A) Strict consensus cladogram of six trees of the first 654 nt at the 5' end of the Cf-2 homologs. (B) Unique phylogram of the first 654 nt at the 5' end of only full-coding Cf-2 homologs; this tree was used in the PAML (1) analysis. (C) Unique phylogram of the 1,044 nt at the 3' end of full-coding Cf-2 homologs; this tree was used in the PAML analysis. (D) Unique phylogram of the 1,491 nt at the 3' end of full-coding Cf-2 homologs; this tree was used in PAML analysis. (E) Strict consensus cladogram of two trees of the 1,045 nt at the 3' end of the Cf-2 homologs.
Table 8. GENECONV (1) results for sequence exchange among haplotypes
|
Sequence set |
Sequence names* |
P value† |
Fragment‡ begins |
Fragment ends |
Length , bp |
No. of polymorphic§ sites |
|
All size classes |
Hcr2-p7 :Hcr2-p8 |
0.0000 |
2295 |
3339 |
1045 |
50 |
|
|
Hcr2-p3 : Hcr2-p4 |
0.0071 |
374 |
711 |
338 |
53 |
|
|
Hcr2-p2 : Hcr2-p6 |
0.0450 |
1 |
380 |
380 |
34 |
|
|
Hcr2-p8 |
0.0151 |
69 |
124 |
56 |
5 |
|
Only full ORFs |
Hcr2-p3 :Hcr2-p4 |
0.0000 |
374 |
719 |
346 |
46 |
|
|
Hcr2-p2 :Hcr2-p4 |
0.0173 |
1849 |
3236 |
1388 |
55 |
|
|
Hcr2-p3 :Hcr2-p5 |
0.0261 |
596 |
719 |
124 |
24 |
|
|
Cf-2 :Hcr2-p4 |
0.0272 |
2647 |
2940 |
294 |
11 |
*Because of low polymorphism within size classes, sequence exchange was considered unlikely and only comparisons among size classes were evaluated.
†
Global P values are Bonferroni-corrected by the program.‡
Sites are numbered according to the Cf-2 size class genes.§
Number of polymorphic sites within the fragment.1. Sawyer, S. A. (1999) GENECONV (Dept. of Mathematics, Washington University, St. Louis).