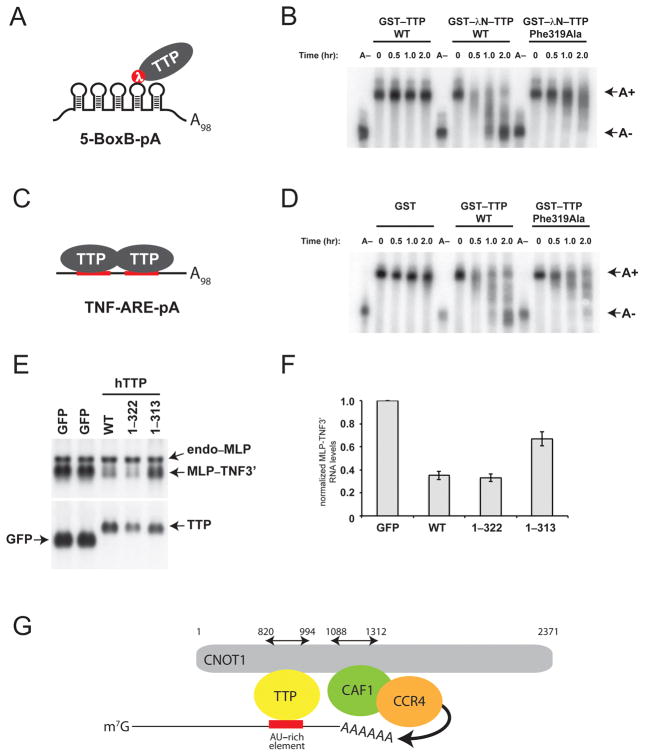
Figure 4.
Disruption of the TTP–CNOT1 interaction impairs mRNA deadenylation in vitro and mRNA stability in vivo. (A) Schematic depiction of GST–λNHA–TTP tethered to 5-BoxB-pA RNA. (B) 5-BoxB-pA RNA deadenylation in Krebs extract in the presence of recombinant wild-type GST–TTP, or wild-type or mutant (Phe319Ala) GST–λNHA–tagged TTP. A(−) RNA was prepared by incubating 5-BoxB-pA with Oligo(dT) and RNase H. Polyadenylated and deadenylated mRNAs are marked on the right of the figure. (C) Schematic depiction of TTP associated with TNF-ARE-pA RNA. (D) TNF-ARE-pA RNA deadenylation in Krebs extract in the presence of recombinant GST, or wild-type or mutant (Phe319Ala) GST–tagged TTP. A(−) RNA was prepared by incubating TNF-ARE-pA with Oligo(dT) and RNase H. Polyadenylated and deadenylated mRNAs are marked on the right of the figure. (E) HEK-293 cells were co-transfected with the reporter construct pMLP-TNF3′ and DNA encoding GFP or TTP protein constructs. Total cellular RNA was harvested and subjected to electrophoresis and Northern blotting. In the upper panel, the Northern blot was probed with a 32P-labeled MLP probe; in the lower panel, the blot was probed with a 32P-labeled TTP probe. In the upper panel, the endogenous MLP mRNA (endo-MLP) the reporter MLP-TNF3′ species are indicated by arrowheads. In the lower panel, the expression levels of transcripts for GFP alone and GFP–TTP are indicated. (F) Quantification of MLP-TNF3′ RNA levels (see Figure 4E), normalized to those in GFP-expressing cells, in the presence of GFP, TTP WT, TTP 1–322 and TTP 1–313. Mean values ±s.e. from seven independent experiments are shown. (G) Model for structural organization of mRNA-bound TTP in complex with CNOT1 and CAF1 and CCR4 deadenylases. The cartoon summarizes structural data reported in this manuscript, combined with data from crystal structures of yeast 20 and human 21 NOT1 proteins in complex with CAF1 and CCR4 deadenylases. TTP and CAF1 binding domains in CNOT1 (820–999 and 1088–1312, respectively) refer to coordinates of human CNOT1 isoform c. Other CCR4–NOT subunits (i.e. CNOT2, 3, 9 and 10) are not shown.