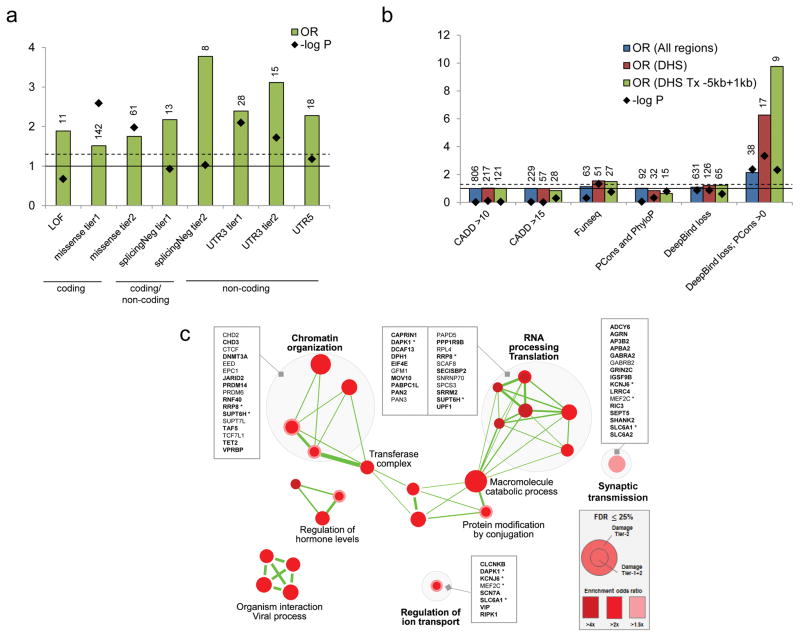
Figure 3.
Functional impact of genome-wide damaging de novo mutations. (a) Damaging de novo mutations are significantly enriched in both coding (missense) and non-coding regions (splicingNeg, UTR3 and UTR5) in ASD compare to population controls. Definition of damaging tiers can be found in the Methods. LOF: loss of function mutations; missense: missense mutations; splicingNeg: exon-skipping mutations predicted by SPANR; UTR: untranslated regions. Number of variants is indicated above each bar. Solid horizontal line indicates OR=1, and dash horizontal line represents p=0.05. (b) Non-coding de novo mutations in non-genic regions are significantly enriched in DNase I hypersensitive regions (DHS). Damaging de novo mutations predicted by “Deepbind loss; PCons >0” are significantly enriched in ASD in general (All regions), but further enriched in DNase I hypersensitive regions and regions proximal to genes. DHS: DNase I hypersensitive sites; Tx: transcript; PCons: PhastCons. Number of variants is indicated above each bar. Solid horizontal line indicates OR=1, and dash horizontal line represents p=0.05. (c) Damaging de novo mutations are significantly enriched (FDR ≤0.25) in Gene Ontology defined pathways that are related to chromatin organization, RNA processing and translation, synaptic transmission, and others. Genes involved in the pathways are listed. Genes with DNMs in coding region are bolded. Asterisk represents gene that is found in more than one gene-pathway.