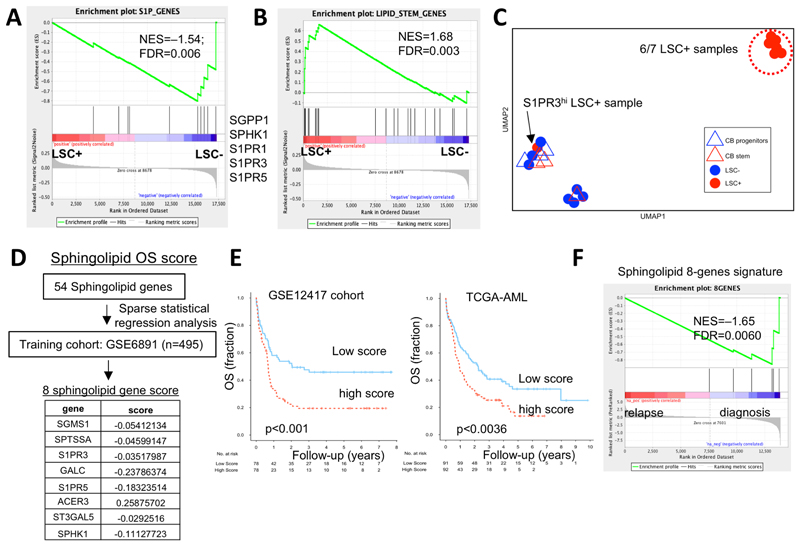
Figure 6. Sphingolipid genes including S1PR3 predict prognosis in human AML.
A, GSEA analysis of 10 S1P genes in AML LSC+ and LSC– gene expression data shows enrichment of S1PR1, S1PR3, S1PR5, the S1P kinase (SPHK1) in LSC– samples. See Supplementary Table 12 for LSC ranking. B, GSEA analysis of 23 lipid-stem genes, previously shown to be enriched in HSC subsets relative to committed progenitor populations, in AML LSC+ and LSC– gene expression data shows enrichment in LSC+ samples. C, A uniform manifold approximation and projection (UMAP) plot for the sphingolipid composition of samples measured by LC/MS spectrometry in functionally defined individual LSC+ (n=7) and LSC–(n=7) fractions from 10 patient AMLs as determined by xenotransplantation and normal stem (CD34+CD38-) and progenitor (CD34+CD38+) fractions (CB, n=3). LSC+ (red circles), LSC– (blue circles), CB stem (red triangles), CB progenitor (blue triangles). LSC+ subpopulation from AML9, which is high for S1PR3 gene expression, clusters away from other 6 LSC+ samples. See Supplementary Figure 9C-G for sphingolipid composition distribution of individual samples and quantification of specific sphingolipid species and Supplementary Table 9 for patient AML data. D, Sparse regression analysis, as described in Ng, et al. al(7), to derive a weighted sphingolipid gene score predictive of overall survival (OS) applied to a large training cohort of n=495 AML patients yielded a set of 8 genes with their associated scores which were then validated on GSE12417 and TCGA-AML cohorts in univariate analysis. These included 3 genes associated with S1P production and signaling and 5 genes encoding sphingolipid enzymes. E, Kaplan-Meier estimates of OS based on above versus below median values of a weighted sphingolipid gene score. A low score is associated with greater OS. F, GSEA plot for sphingolipid 8-gene OS signature shows negative enrichment in patient samples at relapse.