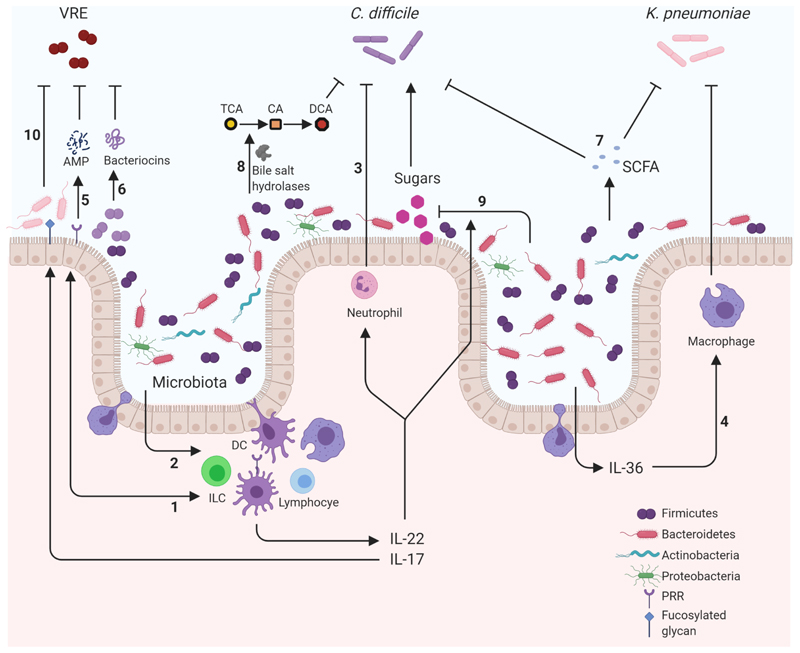
Figure 1.
Mechanisms of microbiota mediated colonization resistance against pathogens within the gastrointestinal tract. Representation of pathways involving components of the immune system and microbiota that help suppress gastrointestinal tract colonization by Clostridioides difficile, Klebsiella pneumoniae and vancomycin-resistant enterococci (VRE). Suppressive mechanisms involve a variety of important pathways that can be broadly classified into four interconnected categories. Cellular interaction: Microbiota activating intestinal epithelia (1) or immune cells (2) induce production of proinflammatory cytokines such as IL-22 and IL-17. These cytokines promote clearance of C. difficile through the regulation of important innate cells such as neutrophils (3). Important phyla such as Bacteroidetes induce the production of IL-36 which promotes macrophage-mediated clearing of K. pneumoniae (4). Antimicrobial production: Pattern recognition receptor (PRR) detection of the microbiota induces the production of antimicrobial peptides (AMP) such as REGIIIγ that is protective against VRE (5). Members of the microbiota can also produce their own AMPS such as bacteriocins. For example, Blautiaproducta produce lantibiotics which inhibit VRE (6). Metabolic: Metabolic products from the microbiota, including short-chain fatty acids (SCFA), can be antagonistic to pathogens reducing the fitness of C. difficile and acidifying K. pneumoniae intracellularly (7). Enzymes produced by the microbiota can also metabolise host compounds into products that are disruptive to pathogens. The bile acid taurocholic acid (TCA) is deconjugated by microbiota bile acid hydrolases into cholic acid (CA) and subsequently into deoxycholic acid (DCA) which is inhibitory to C. difficile growth (8). Nutritional immunity: Nutrients are limited resources and so utilisation by the microbiota diminishes availability to incoming pathogens. IL-22-induced N-glycosylation promotes a microbiota that utilise sialic acid and succinate reducing their abundance preventing the expansion of C. difficile (9). Similarly, IL-22 induction of glycan fucosylation promotes anaerobic commensals competing with VRE limiting its expansion (10). These examples demonstrate how the microbiota provide resistance to three pathogens by engaging with a multitude of mechanisms that are antagonistic to the success of the pathogens in the gastrointestinal tract. DC: Dendritic Cell, ILC: Innate Lymphoid Cell. Created with BioRender.com.