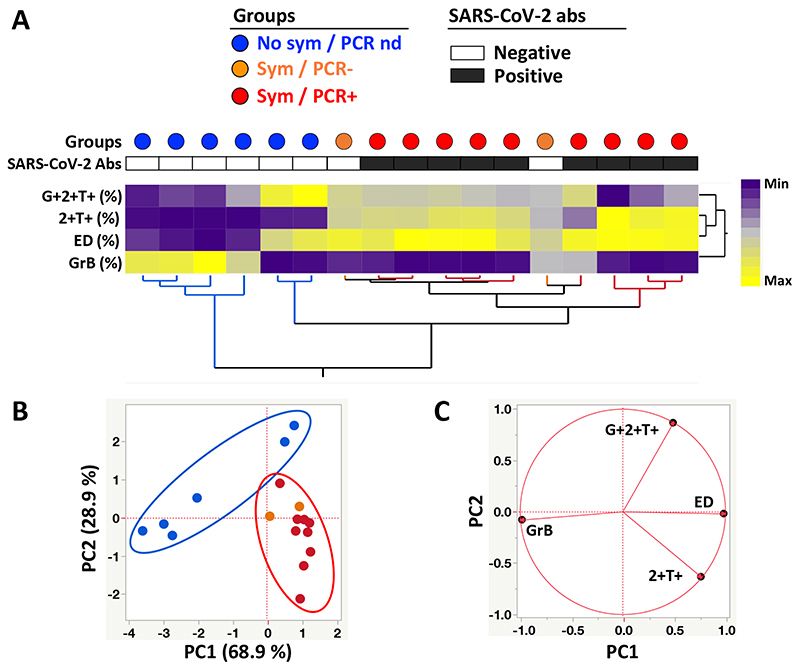
Figure 5. Phenotypic signature of SARS-CoV-2-specific IFNγ+ CD4+ T cells according to clinical characteristics.
A- Non-supervised two-way hierarchical cluster analysis (HCA, Ward method) using three phenotypic parameters (i.e. GrB expression and the proportion of ED and IFNγ+TNFα+IL2-) from SARS-CoV-2-specific CD4 T cells. Each column represents a participant and is color-coded according to their clinical characteristics indicated by a dot at the top of the dendrogram (Blue: no symptoms, no SARS-CoV-2 PCR performed; Orange: self-reported symptoms, SARS-CoV-2 PCR negative; and Red: self-reported symptoms, SARS-CoV-2 PCR positive). Participants with a positive SARS-CoV-2 serology test are indicated by a black box and a white box identifies SARS-CoV-2 antibody-negative subjects. Data are depicted as a heatmap coloured from minimum to maximum values for each parameter. B- Principal component analysis on correlations, derived from the three studied parameters. Each dot represents a participant. The two axes represent principal components 1 (PC1) and 2 (PC2). Their contribution to the total data variance is shown as a percentage. C- Loading plot showing how each parameter influences PC1 and PC2 values.