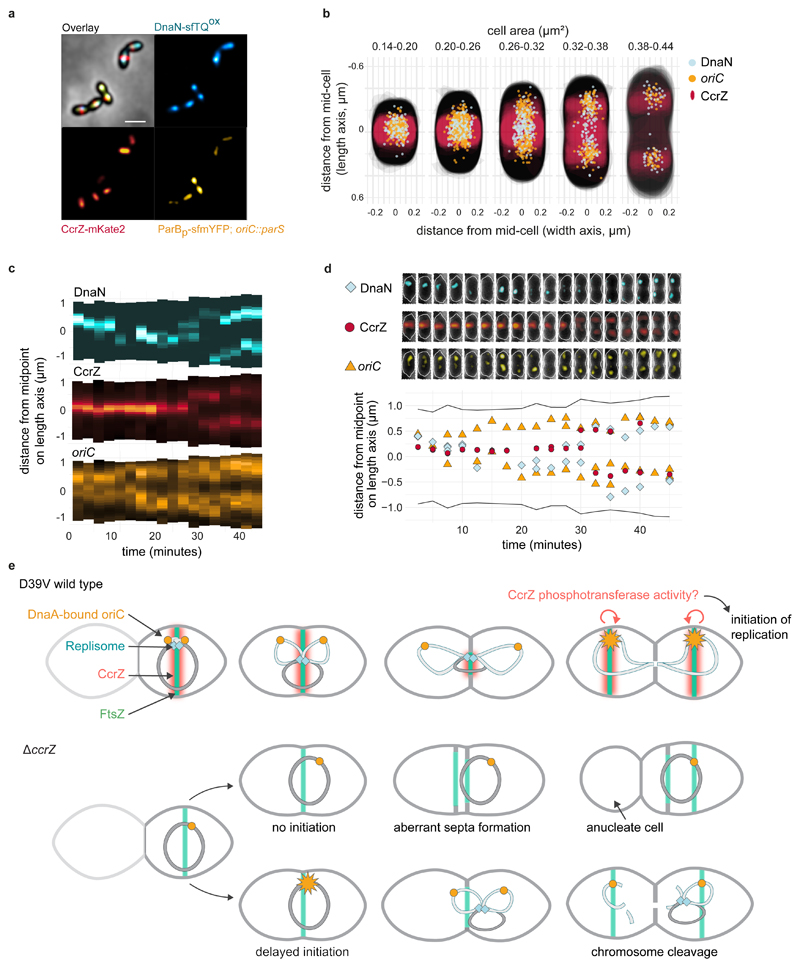
Fig. 6. Spatio-temporal localization of CcrZ via FtsZ ensures proper timing of DNA replication in S. pneumoniae .
a, Microscopy of the origin of replication (yellow), replication fork (cyan) and CcrZ (red) in live S. pneumoniae wild type background cells. Scale bar, 1 μm. b, DnaN, oriC and CcrZ localizations grouped by cell area (μm2) in five equally sized groups. Analyzed from snapshots of exponentially growing cells. c, Single-cell kymographs of DnaN, CcrZ and oriC localizations in a 2:30 minute interval time-lapse movie. d, Tracked DnaN, oriC and CcrZ over time in a single cell. Top: overlay of fluorescence, cell outline and phase-contrast of the cell displayed in panel c and bottom: fluorescence localization on the length axis of the same single cell over time. e, Model for spatio-temporal control of replication by CcrZ. In S. pneumoniae, CcrZ is brought to the middle of the cell where the DnaA-bound origin of replication is already positioned. CcrZ then stimulates DnaA to trigger DNA replication by an as of yet unknown activity, possibly involving a phosphor-transfer event. While the precise regulation and localization of CcrZ seems diverse between different organisms, CcrZ’s activity to stimulate DNA replication is conserved, at least in S. pneumoniae, S. aureus and B. subtilis.