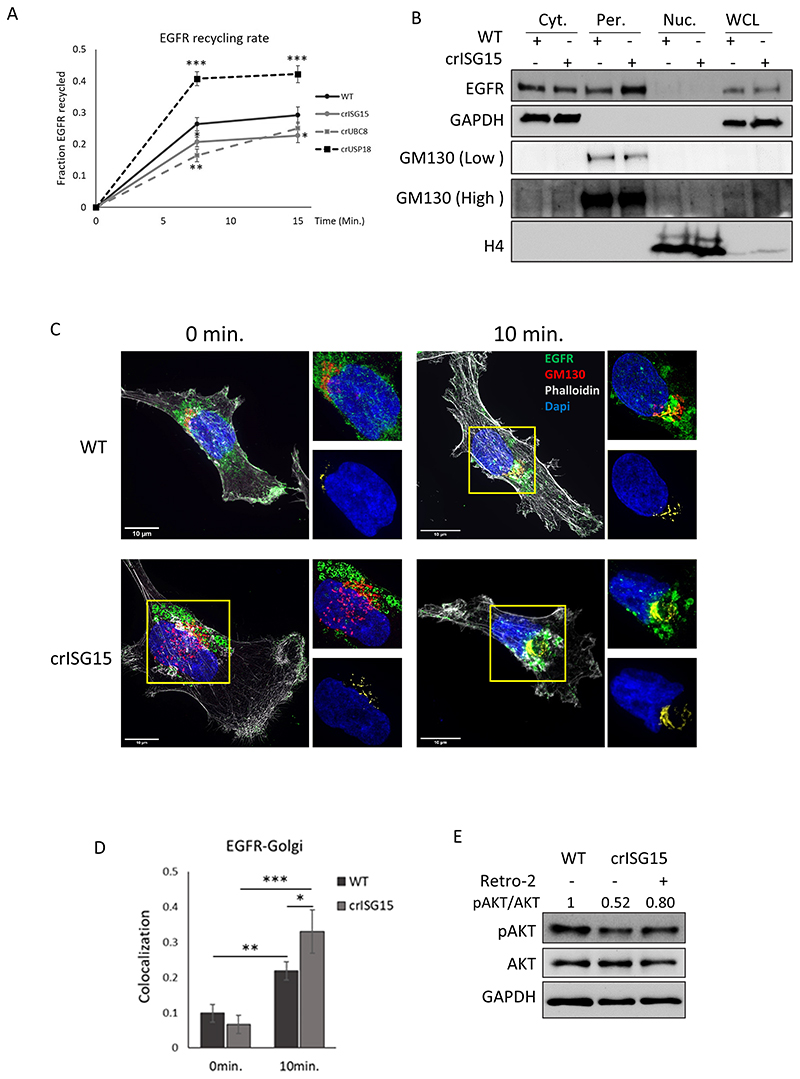
Fig. 4. ISGylation promotes faster EGFR recycling.
(A) Graph showing EGFR membrane recycling rate. Cells were surface labelled with NHS-SS-biotin, and surface receptors allowed to internalise for 30 mins in serum free medium. Biotin remaining at the cell surface was removed, and internalised receptors allowed to recycle to the plasma membrane for the indicated time. Biotin label was removed from surface proteins at the cell membrane, cells were lysed and the fraction of recycled EGFR was determined at each timepoint. p-value < 0.05 (*), p-value < 0.01 (**), p-value < 0.005 (***). (B) Analysis by WB of EGFR localization in WT and crISG15 cells stimulated with EGF for 10min. Cells were fractionated into membrane and cytoplasm fraction (Cyt.), perinuclear region (Per.) and nucleus (Nuc.). EGFR levels detected by WB in the different fraction and the presence of the respective markers of subcellular localisation, GAPDH (cytoplasm), GM130 (Golgi) and Histone 4 (H4; Nucleus). Whole cell lysates (WCL) were included as reference of relative protein expression. (C) Representative super-resolution confocal images, obtained at 100x, of WT and crISG15 cells pre-treated with cycloheximide for 15 min. and stimulated for 10 min. with EGF 10 ng/ml. Cells were fixed, permeabilised and incubated with antibodies as indicated. Images show EGFR in green, GM130 as Golgi marker in red, phalloidin in grey and DAPI, in blue. At top right of each image, zoom of Golgi structure displayed. At bottom right of each image, visualisation of the co-localization between EGFR and the Golgi marker GM130 of the images in yellow and DAPI, in blue as reference. 10μm scale bars are displayed in the bottom-left corner. (D) Bar graph shows the average EGFR-GM130 co-localization showed in (C) using Costes method, average ± SD; n=8 field of view. p-value < 0.05 (*) (E) Endosome to Golgi trafficking inhibition induces pAkt levels in crISG15 cells. WT and crISG15 cells treated for 10 min. with EGF. crISG15 cells were pre-treated with retro-2 for 15min or not. Quantification of Akt activation, measured as pSer473-Akt/Akt signal, normalized to WT.