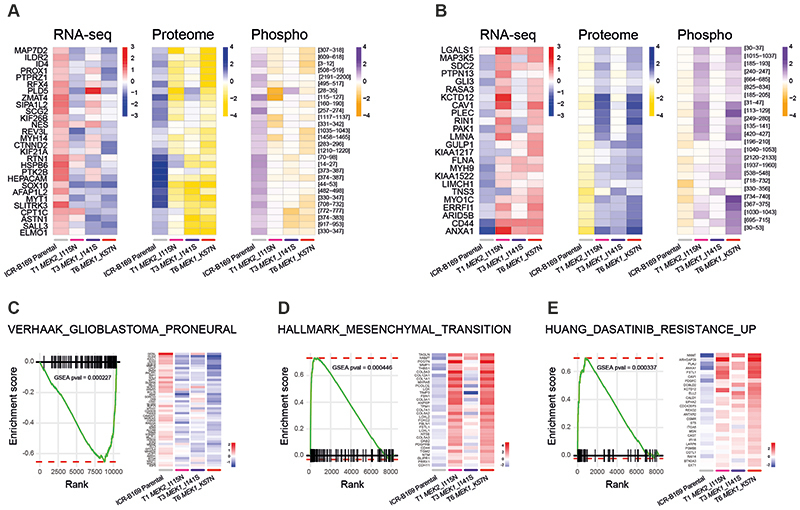
Figure 4. Integrated gene and protein expression profiling of trametinib-resistant DIPG cells.
(A) Co-ordinately down-regulated genes (left), proteins (middle) and phospho-sites (right) in all three trametinib-resistant subclones of ICR-B169 BRAF_G469V cells, as compared to parental. (B) Co-ordinately up-regulated genes (left), proteins (middle) and phospho-sites (right) in all three trametinib-resistant subclones, as compared to ICR-B169 parental (grey). T1 MEK2_I115N, pink; T3 MEK1_I141S, purple; T6, MEK1_K57N, red. (C) Gene set enrichment analysis (GSEA) enrichment plots for the signature VERHAAK_GLIOBLASTOMA_PRONEURAL in T6 MEK1_K57N cells compared to ICR-B169 parental. The curves show the enrichment score on the y axis and the rank list metric on the x axis. Alongside is a heatmap representation of expression of significantly differentially expressed genes in the signature in all three trametinib-resistant clones compared to parental. (D) GSEA enrichment plots for the signature HALLMARK_EPITHELIAL_MESENCHYMAL_ TRANSITION in T6 MEK1_K57N cells compared to ICR-B169 parental. The curves show the enrichment score on the y axis and the rank list metric on the x axis. Alongside is a heatmap representation of expression of significantly differentially expressed genes in the signature in all three trametinib-resistant clones compared to parental. (E) GSEA enrichment plots for the signature HUANG_DASATINIB_RESISTANCE_UP in T6 MEK1_K57N cells compared to ICR-B169 parental. The curves show the enrichment score on the y axis and the rank list metric on the x axis. Alongside is a heatmap representation of expression of significantly differentially expressed genes in the signature in all three trametinib-resistant clones compared to parental. T1 MEK2_I115N, pink; T3 MEK1_I141S, purple; T6, MEK1_K57N, red.