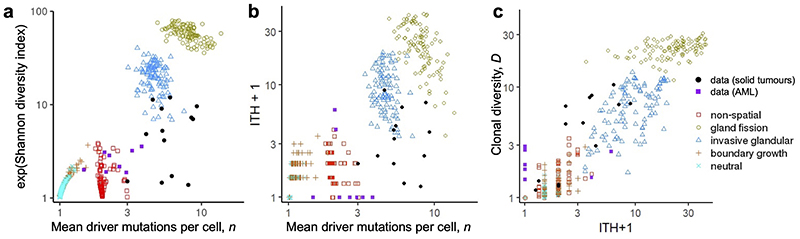
Extended Data Fig. 9. Alternative diversity indices.
a, Variation in the exponential of the Shannon diversity index versus mean number of driver mutations per cell (n). b, Variation in the ITH index versus mean number of driver mutations per cell (n). c, Correlation between the inverse Simpson index (D) and the ITH index. Coloured points correspond to four example models with different spatial structures and different manners of cell dispersal but identical driver mutation rates and identical driver mutation effects (100 stochastic simulations per model). Neutral counterparts of the four models are represented together as an additional group. Mutations with frequency less than 1% are removed from model outcomes before calculating ITH and D. Black circular points show values derived from multi-region sequencing of kidney cancers, lung cancers and breast cancers. Purple squares show values derived from single-cell sequencing data for acute myeloid leukaemia. Estimates of the Shannon index and ITH index based on multi-region sequencing data are expected to be lower than true values because these indices are sensitive to the removal of rare types, many of which are likely to be missing from the data due to sampling error.