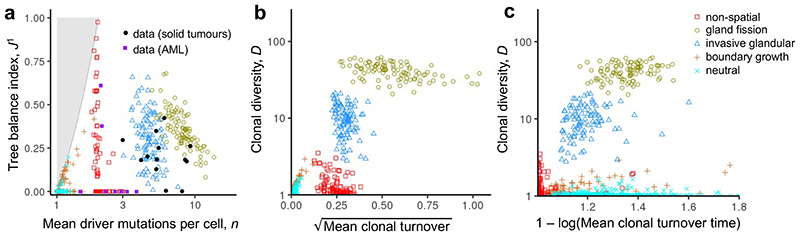
Figure 4. Alternative summary indices for characterizing modes of tumour evolution.
Index values are shown for four models representative of their evolutionary modes with different spatial structures and different manners of cell dispersal but identical driver mutation rates and identical driver mutation effects (100 stochastic simulations per model). Neutral counterparts of the four models are represented together as an additional group. a, Tree balance J 1 versus mean number of driver mutations per cell n. Black circular points show evolutionary indices derived from multi-region sequencing data. Solid purple squares show values derived from single-cell sequencing data for acute myeloid leukaemia. Coloured hollow points are the predictions of the four models and their neutral counterparts, excluding mutations with frequency below 1%. It is impossible to construct trees for locations within the shaded region. b, Clonal diversity D versus mean clonal turnover . c, Clonal diversity D versus mean clonal turnover time . Both the mean clonal turnover index and the mean clonal turnover time have been transformed to equalize cluster widths and facilitate comparison between plots. In panel c, low x-axis values indicate that most clonal turnover occurred late in tumour growth.