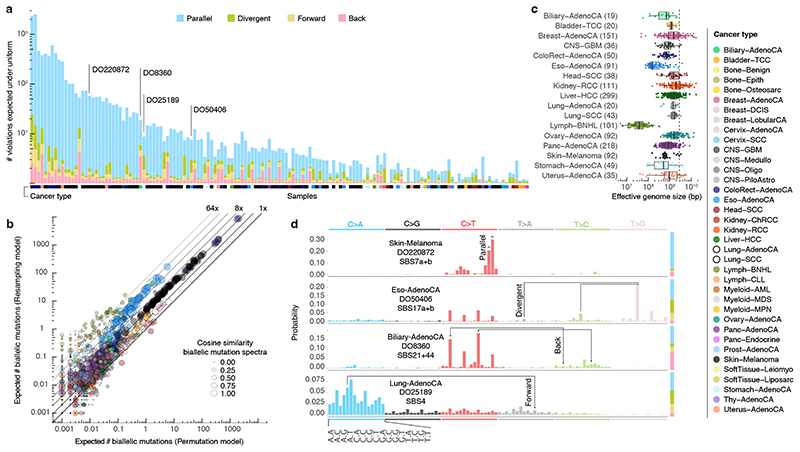
Figure 2. Simulated landscape of infinite sites violations in the PCAWG cohort.
(a) Number and type of infinite sites violations in 147 PCAWG samples with ≥ 1 expected violation under a uniform mutation distribution. Bar height indicates the expected number of violations and coloured subdivisions represent the fractions contributed by each violation type. Tumour type of the samples is colour-coded below the bars. The four samples highlighted in (d) are indicated. (b) Comparison of the expected biallelic violations from the uniform permutation and neighbour resampling models. Every dot represents a tumour simulated 1,000x with each model. Colour and size reflect, respectively, tumour type and the cosine similarity of the predicted biallelic mutation spectra. (C) Box and scatterplot showing the effective genome size perceived by the mutational processes per cancer type, as estimated from the per-sample differences between simulation approaches. The dashed line indicates the callable genome size. The effective genome size is smallest in Lymph-BNHL (~37Mb), likely driven by recurrent focal hypermutation 13 . Centre line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. Only tumours with ≥ 10 biallelic mutations across 1,000 simulations are included and their numbers are indicated between parentheses next to the tumour type. Only tumour types with ≥ 10 such tumours are shown. (d) Mutation spectra of four tumours with distinct violation contributions indicated in (a). The 16 distinct trinucleotide contexts are provided on the x-axis for C>A type substitutions and are the same for each coloured block. The proportion of parallel, divergent, back and forward mutation is indicated in the stacked bar on the right. Frequent combinations of mutations leading to specific infinite site violations are highlighted as well as the signatures generating them.