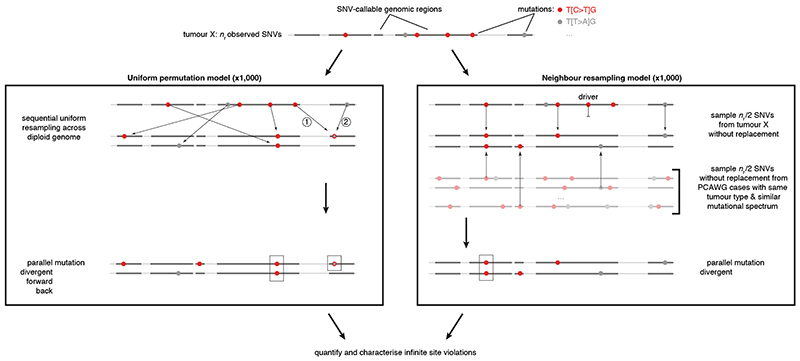
Extended Data Fig. 1. Simulation approaches for infinite sites violations.
Schematic overview of the uniform permutation (left) and neighbour resampling (right) approaches to assess the number and type of infinite sites violations expected in a tumour. Numbers in the uniform permutation panel highlight the sequential nature of the sampling, which keeps track of mutated positions to consider them accordingly for biallelic, forward, and back mutation. Note that the neighbour resampling model excludes all PCAWG annotated driver mutations and allows simulation of biallelic events only.