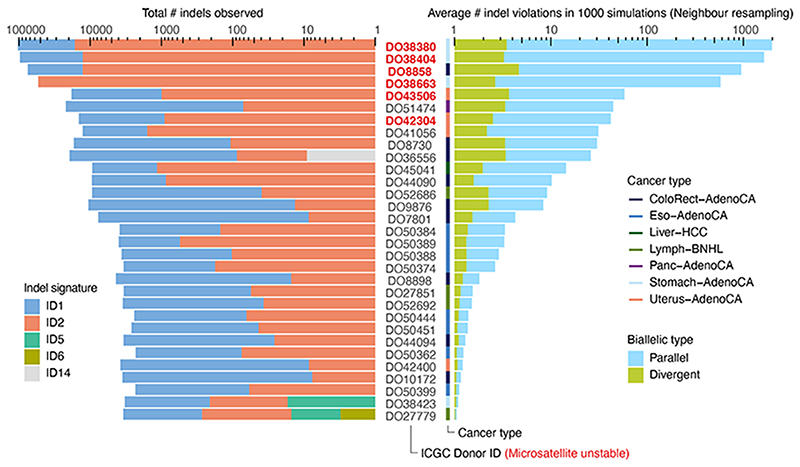
Extended Data Fig. 2. Biallelic indels are expected in a subset of microsatellite unstable tumours.
Bar plots of the observed indel burden and signature (left) and the expected biallelic indels according to the neighbour resampling model (right). Bar height indicates total numbers and coloured subdivisions represent fractions contributed by each indel signature (left) or biallelic indel type (right). Only PCAWG tumours with ≥1 expected biallelic indel are shown. Four microsatellite unstable tumours are predicted to boast several hundreds to over one thousand, mostly parallel, biallelic indels. These mainly originate from indel signatures 1 and 2, likely reflecting slippage during DNA replication and subsequent 1bp T (or A) insertion and deletion in thymine (adenosine) mononucleotide repeats, respectively.