Abstract
Experimental mouse models are central to basic biomedical research; however, variability exists across genetically identical mice and mouse facilities making comparisons difficult. Whether specific indigenous gut bacteria drive immunophenotypic variability in mouse models of human disease remains poorly understood. We performed a large-scale experiment using 579 genetically identical laboratory mice from a single animal facility designed to identify the causes of disease variability in the widely used dextran sulphate sodium (DSS) mouse model of inflammatory bowel disease. Commonly used treatment endpoint measures, weight loss and intestinal pathology, showed limited correlation and varied across mouse lineages. Analysis of the gut microbiome, coupled with machine learning and targeted anaerobic culturing, identified and isolated two previously undescribed species, Duncaniella muricolitica and Alistipes okayasuensis, and demonstrated that they exert dominant effects in the DSS model leading to variable treatment endpoint measures. We show that the identified gut microbial species are common, but not ubiquitous, in mouse facilities around the world, and suggest that researchers monitor for these species to provide experimental design opportunities for improved mouse models of human intestinal diseases.
Experimental mouse models are central to basic biomedical research and serve as important pre-clinical models in drug discovery for a variety of human diseases, including autoimmune and metabolic disorders, cancers and infections. Unfortunately, there is tremendous variability in disease penetrance and reproducibility among genetically identical mice within and between mouse facilities, limiting the utility of many commonly used models 1–3 . Specific pathogen free (SPF) protocols have been successfully employed to control the confounding effects that can be caused by known bacterial pathogens, but we lack equivalent knowledge of the effects of the diverse, uncharacterized symbiotic bacteria within the indigenous gut microbiota of mice. While our knowledge of the human gut microbiota in health and disease has expanded extensively with large-scale microbiome studies, comprehensive computational bacterial discovery 4,5 , and extensive genome sequenced bacterial collections 6–9 , equivalent investigations in mouse models has remained limited. The vast differences in species type, composition and dispersal between human and mouse microbiotas highlight the importance of efforts to culture mouse specific bacteria and to validate their functions during intestinal homeostasis and disease 10,11 .
Inflammatory bowel disease (IBD) research relies heavily on laboratory mice to understand colitis and the role of host genetics in disease resistance and susceptibility. Detailed immune characterisation shows colitis induction with specific microbial communities 12 or species colonisation with Helicobacter species 13,14 , Enterobacteriaceae 15 , Bilophila wadsworthia 16 , or segmented filamentous bacteria 17 . Potential genetic modulators of IBD are routinely assessed through administration of Dextran Sulphate Sodium (DSS) at various concentrations in drinking water, which acts by damaging the gut epithelial barrier to trigger inflammatory disease 12,13 . In germ free mice, immune mediated colitis is largely absent, though reduced barrier function can introduce susceptibility at higher DSS concentrations 18 . Similarly, dietary changes in both germ free and SPF mice can determine DSS susceptibility in a microbiome dependent and independent manner 19 , and highly specific protection is provided by the presence of Clostridium immunis 20 . Despite these advances, the gut microbiota in these mouse models, including standard SPF models, is rarely profiled at a genomic level or cultured in the laboratory, nor robustly controlled to understand the contribution of the general gut bacteria to experimental outcomes.
To quantify the variability of the widely used DSS model of IBD and to investigate primary factors contributing to this variability, we undertook a large-scale experiment using 579 genetically identical laboratory mice in a single animal facility. Incorporating standardized phenotyping, microbiome analysis and metadata collection, this approach was designed to identify and validate the causes of variability for standard experimental endpoints (weight loss and intestinal pathology). Using a data-driven approach, we identified from gut microbiome data, candidate bacterial taxa driving weight loss and intestinal inflammation. Functional validation with cultured isolates suggests these specific, bacterial strains can determine experimental outcomes in gnotobiotic mice. Thus, using an experimental approach for microbiome discovery, we identified the previously undescribed mouse gut bacteria, Duncaniella muricolitica and Alistipes okayasuensis, found frequently in mouse colonies around the world, that impact outcomes in the DSS model of colitis and may be acting as pathobionts.
Results
Disease severity is highly variable in the DSS mouse model
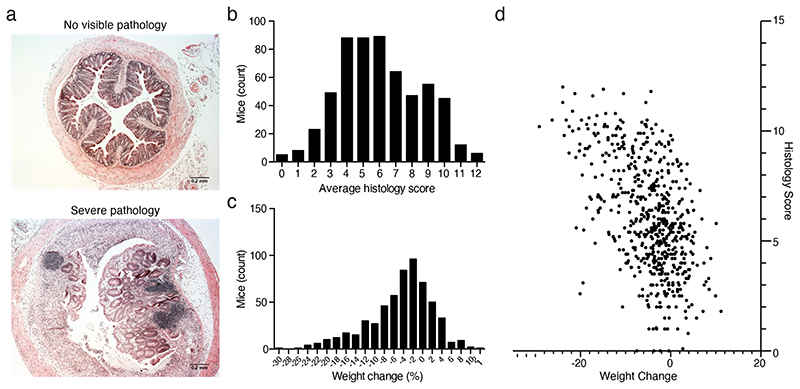
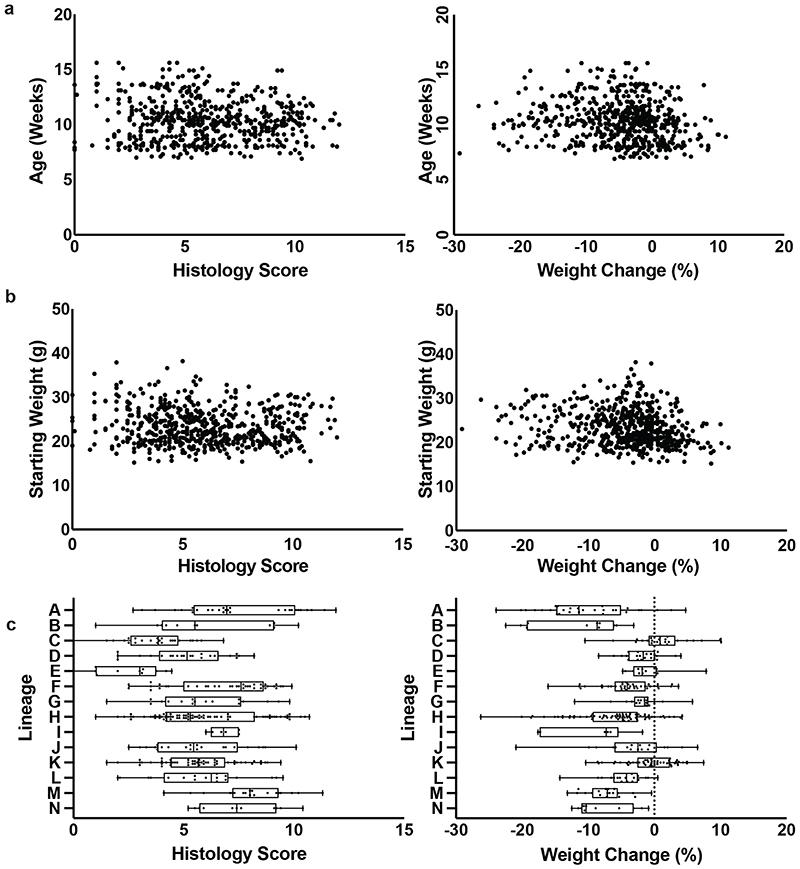
We administered 1.5% DSS through the drinking water for 7 days to 579 identically housed, SPF wild type C57BL/6N mice (297 males, 282 females; median 11 weeks; SD 12 days), from 14 distinct parental lineages (i.e. founder matings derived from a single breeding pair). We observed extensive variability in experimental endpoints in these genetically identical mice at 10 days post DSS treatment, with histological assessment of intestinal pathology of the mid and distal colon ranging from severe inflammation, characterised by extensive crypt loss, leukocytic infiltration and edema, to no visible inflammation (Fig. 1a, 1b). Substantial variation was also observed in the weight change of mice treated with DSS, with the response ranging from a 12% weight gain to a 30% weight loss across the cohort (Fig. 1c). Regression analysis considering histology scores and weight loss demonstrated limited correlation (R2 = 0.27) suggesting that weight loss and intestinal pathology outcomes, common experimental endpoint measures in the DSS model, may be partially-independent responses to DSS (Fig. 1d). This relationship is consistent with systemic complications contributing to experimental endpoints in the DSS model.
Figure 1. Disease severity is highly variable in the DSS mouse model.
(a) Representative images of extremes of mid-colon inflammatory response of wild-type C57/BL6 mice after treatment with DSS showing a score of 0 and a score of 12. Distribution of (b) average histology scores and (c) weight change across 579 (297 males, 282 females; median 11 weeks; SD 12 days) wild-type C57BL/6N mice exposed to DSS treatment. (d) Relationship between weight change and histology score across 579 (297 males, 282 females; median 11 weeks; SD 12 days) R2 correlation of 0.273.
Gut microbiota is major driver of variable response to DSS
To identify the primary factors determining disease outcome, we established a random forest classifier model incorporating mouse sex, age, lineage, parents, mouse room and experimental date to assess the relative importance of each of these factors (Extended Data Fig. 1). The strongest predictive factors for weight loss and histology score, both together and independently in these genetically identical C57BL/6N mice, maintained under standardized SPF conditions within the same facility, was the parental lineage, with correlation analysis confirming these associations (p < 0.01; Kruskal-Wallis; Extended Data Fig. 1).
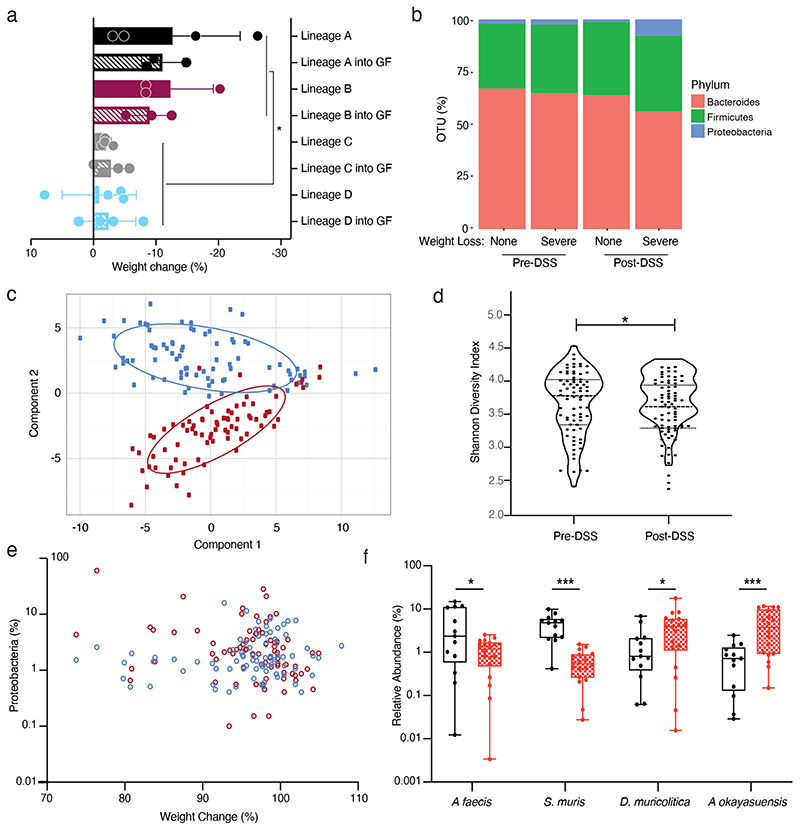
Since all the mice were derived from the same genetic founder stock the most likely mediator of their DSS phenotype was an extrinsic lineage specific factor. However, to exclude the role of host genetics or epigenetic differences between colonies, and to determine if the microbiota alone could be responsible for the observed phenotypic variation, faecal samples were collected from SPF mice (without DSS treatment) from parental lineages that represented extremes of disease outcome and used to colonise C57BL/6N germ-free mice. These colonised recipient mice were then subjected to DSS treatment with daily weight measurements with colon sections collected and analysed histologically 10 days after the initiation of DSS treatment. The recipient mice exhibited both weight loss and pathology scores that were not statistically different from those observed in the respective donor mouse lineages but differed significantly between disease-associated and non-associated lineages (p = 0.0002; paired t-test) (Fig. 2a). From these results it is clear that, independent of other factors such as host genetics or epigenetic lineage specific factors, the microbiota is sufficient to produce the observed variation in phenotypic response to DSS administration.
Figure 2. Discovery of candidate bacterial taxa associated with DSS disease variability.
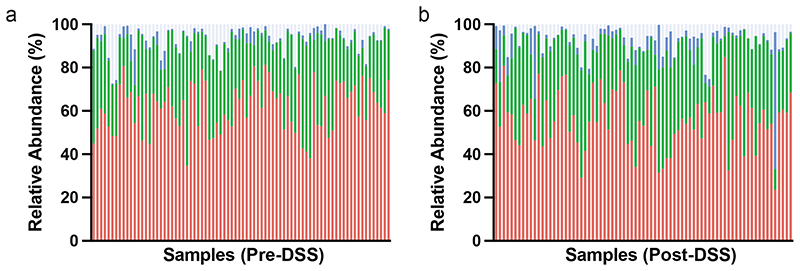
(a) Comparison of weight changed observed in conventional SPF mice (Lineage A, B, C and D; solid) and germ-free mice colonized by oral gavage (Lineage A, B, C and D into GF; hatched) after 10 day DSS challenge (p = 0.0002 two tailed T-test with Welch's correction, Data plotted as mean +/-SD. n = 4 (Lineage A, Lineage C, Lineage C + GF, Lineage D and Lineage D + GF) and n = 3 (Lineage B, Lineage B+GF, Lineage A+GF) animals). (b) Distribution of Bacteroides (red), Firmicutes (green) and Proteobacteria (blue) in pre- and post-DSS samples that exhibited asymptomatic or greater than 5% weight loss and inflammation. (c) Principal component analysis of the 16S rRNA amplicon sequencing profiling identifies a clear change between pre-DSS samples (blue) and post-DSS samples (red) regardless of lineage (d) Alpha diversity using Shannon-diversity index grouped by Pre-DSS and Post-DSS samples (* p = 0.0357; paired T-test, n=77). (e) Relationship between weight change and relative abundance of Proteobacteria pre-DSS challenge (blue; Spearman Correlation 0.196) and post-DSS (red; Spearman Correlation -0.289). (f) Relative abundance of A faecis, A okayasuensis, D muricolitica and S. muris prior to challenge with DSS amongst samples from mice that experience greater than 5% weight loss (red) and no weight loss (black) (p = 0.0264, 0.0006, 0.0351, 0.0010; two tailed T-test, n=18, 13 independent animals. Minima, 25th percentile, Median, 75th percentile and maxima are shown).
Discovery of bacteria that drive disease variability
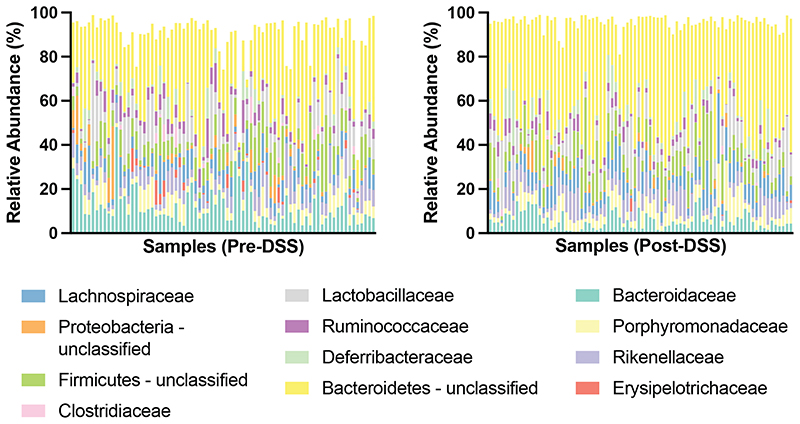
Having established the importance of the microbiota in mediating phenotypic outcomes to DSS in genetically identical mice, we next sought to understand the degree of variability in mouse microbiota across a single facility and define the changes in the microbiota that occur with DSS treatment. Faecal samples were collected prior to DSS treatment and 10 days after DSS exposure, and subjected to taxonomic profiling with 16S rRNA amplicon sequencing to determine the microbiota community composition. Consistent with previous reports of gastrointestinal inflammation, this analysis demonstrated a clear separation of microbiome community between the pre- and post-DSS samples (Fig. 2b, 2c, Extended Data Fig. 2, Extended Data Fig. 3) and a significant reduction in bacterial diversity (Shannon diversity index; p = 0.0357; Fig 2d) 12 . Expansion of Proteobacteria is often associated with gut microbiome dysbiosis, however, while an increase in Proteobacteria was observed post-DSS (p < 0.05, KS), limited association was observed between weight loss and Proteobacteria proportion either pre- (Fig 2e; Spearman Correlation = 0.196) or post DSS treatment (Fig 2e; Spearman Correlation = -0.289). This suggests other gut bacteria may be contributing to the observed variation in DSS resistance and susceptibility respectively.
Given the transmissibility of disease phenotype we observed in the germ-free mouse experiments, we reasoned the observed phenotypic variation was primarily dependent on the baseline microbiota composition, prior to DSS treatment. To understand the composition of the microbiota that was associated with increased risk of disease outcome we applied linear discriminant analysis to the gut microbiota composition prior to and after DSS exposure 24 . This unsupervised analysis leveraged the variability between mouse lineage-specific bacteria to identify the bacterial taxa that most closely predict DSS response. Using this approach, we identified two statistically different bacterial taxa associated with no disease and five statistically different bacterial taxa associated with greater than 5% weight loss. Notably, no relationship was observed between the occurrence or abundance of these isolates in mice pre-DSS exposure and equivalent predictive bacterial taxa were not identified within the microbiota of mice post-DSS exposure. Our results suggest that the presence of specific gut bacterial taxa prior to DSS exposure may influence disease severity and outcome, possibly by either priming or protecting from disease.
Isolation of bacterial species for functional validation
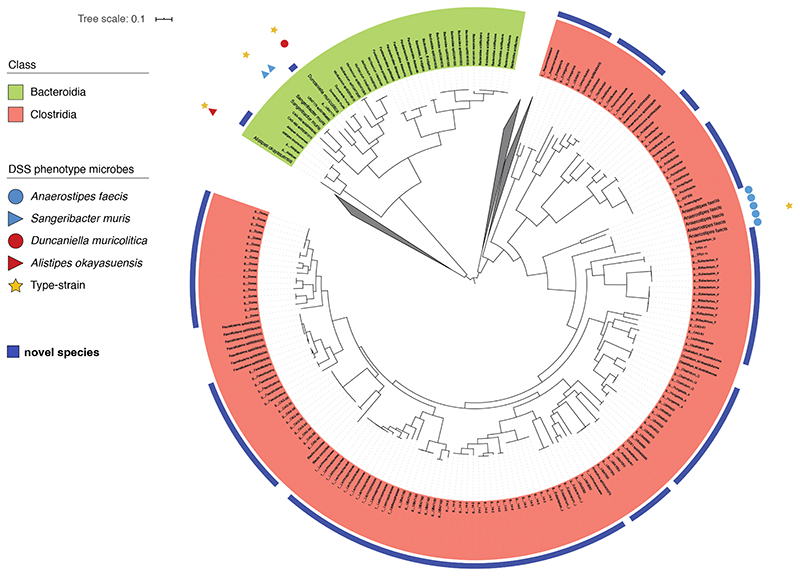
While we show clear associations between specific bacterial taxa and DSS-mediated disease outcome using 16S rRNA gene sequencing, accurate taxonomic identification to strain level and experimental validation requires the isolation and genome sequencing of pure clonal cultures of bacterial strains. To target and purify the bacterial strains, we next performed broad culturing of ~2000 bacterial colonies from mouse faecal samples. The resulting isolates were identified by capillary 16S rRNA gene sequencing, and included four of the previously identified bacterial taxa, 2 “disease-associated” and 2 “health-associated”. These isolates were subjected to whole genome sequencing and genomic analysis to aid in precise taxonomic and phylogenetic assignment (Extended Data Fig. 4). In addition, we performed extensive phenotypic characterization and comparative genome analyses of these 4 species (see Methods) as a basis to provisionally name the health-associated bacteria Anaerostipes faecis and Sangeribacter muris and the disease-associated bacteria Duncaniella muricolitica and Alistipes okayasuensis. Of note, the latter 3 of these species are mouse-specific. Examination of relative abundance of each of these species prior to DSS treatment identified significantly higher levels of A. faecis (p = 0264; T-test) and S. muris (p = 0.0010; T-test) in those mice without weight loss and significantly higher levels of D. muricolitica (p = 0.0351; T-test) and A. okayasuensis (p = 0.0006; T-test) in those mice that experience greater than 5% weight loss (Fig. 2f).
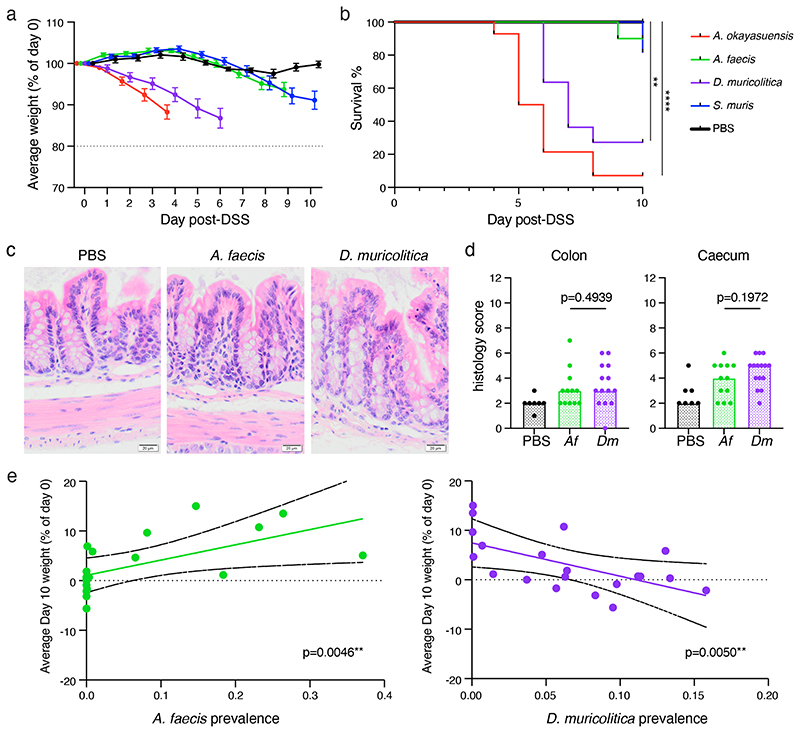
We next repeated the DSS experimental model in germ-free mice or germ-free mice stably mono-colonised for four weeks with one of these four bacterial strains (Extended Data Fig. 5a, Extended Data 6). Control germ-free mice treated with DSS did not lose weight (Fig. 3a) nor experience mortality (Fig. 3b) over the course of the experiment, suggesting that the presence of gut bacteria is important to trigger overt disease in our facility. Comparable experimental outcomes were observed in DSS-treated germ-free mice mono-colonised with either of the health-associated bacteria, which did not lose weight or develop intestinal pathology (Fig. 3a), nor experience significant mortality (Fig. 3b). In contrast, when we DSS treated mice mono-colonised with either of the disease-associated bacteria we observed more rapid and greater weight loss (Fig. 3a) and significantly reduced survival (Fig. 3b). Therefore, we identified and validated D. muricolitica and A. okayasuensis as mouse bacteria that can promote disease not observed in mice mono-colonized with the commensal bacteria S. muris and A. faecis.
Figure 3. Phenotypic analysis of mice mono-colonised with candidate bacterial taxa.
(a) Average weight loss and (b) survival in gnotobiotic mice pre-colonised with either A. okayasuensis (red; n=14 animals; p = 0.00001), A faecis (green; n=10 animals; p = 0.31731), D. muricolitica (purple; n=10 animals; p = 0.00074) or S. muris (blue; n=11 animals; p = 0.16653) and germ-free controls (PBS; black; n=10 animals) in response to oral DSS challenge. All survival curves were compared to PBS control by log-rank (Mantel-Cox) test with median and IQR are plotted. (c) Representative colon images (400X) from two experiments with 3-7 mice per group per experiment and (d) histological scores from the H & E staining of mid-colon (p = 0.4939; Mann Whitney test) and caecum (p = 0.1972; Mann Whitney test) of control germ-free mice (PBS) or mice mono-colonised with A. faecis (n=12) or D. muricolitica (n=14) and challenged with oral DSS for 7 days. Average weight loss curves were censored after the first mouse in each group reached 80% of baseline weight. (e) Day 10 weight loss relative to day 0 starting weight in 20 mice showing a positive correlation (green line) between weight change and prevalence in A. faecis (R2 0.3679; p = 0.0046) and negative correlation (purple line) between weight change and prevalence in D muricolitica (R2 0.3626; p = 0.005) Solid black lines indicate 95% CI.
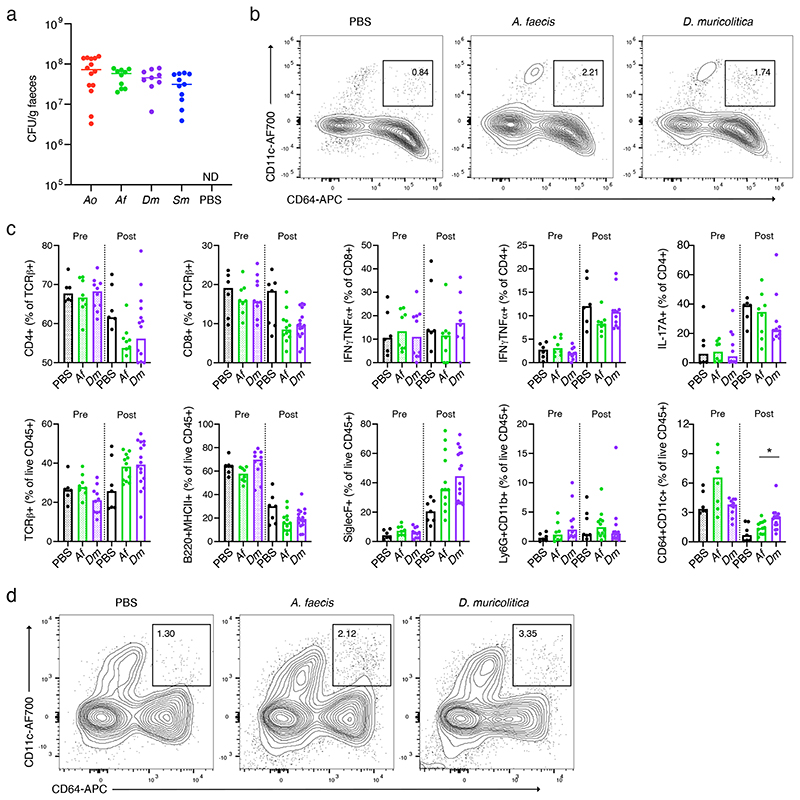
To understand the inflammatory immune responses in the intestines potentially caused by disease-associated versus health-associated species, we next examined histology, immune cell populations and cytokine production. The human colonising A. faecis and the facility prevalent D. muricolitica were chosen as exemplary disease- and health-associated bacteria, respectively, for these analyses. Flow cytometric analysis of intestinal lamina propria leukocytes isolated from germ-free mice mono-colonised for 4 weeks with either A. faecis or D. muricolitica revealed that colonisation with either strain did not significantly impact the composition of intestinal leukocyte and lymphocyte subsets (Extended Data Fig. 5b, c, Extended Data 6). However, the perturbation and epithelial disruption induced by DSS led to significantly increased inflammatory CD64+CD11c+ monocyte/macrophages in the lamina propria compartment of the large intestine in D. muricolitica mono-colonised mice, but not in A. faecis mono-colonised mice (Extended Data Fig. 5c, d, Extended Data 6). These monocytes have been shown to play a pathogenic role in the intestines in the context of Helicobacter-induced inflammation 25 . This suggests that, although not pathogenic or immunogenic at homeostasis, D. muricolitica drives increased inflammatory responses in the wake of epithelial damage whereas health-associated A. faecis does not. Importantly, while histological samples taken after 7 days of DSS treatment from colons and caeca of D. muricolitica mono-colonised mice occasionally exhibited features of severe inflammation, this was not statistically different from that of A. faecis mono-colonised mice (Fig. 3c, d) and did not consistently correspond with the weight loss observed. This again suggests that weight loss in the DSS model may not always correlate with intestinal inflammation and pathology.
To determine if the presence of D. muricolitica or A. faecis had a positive or negative correlation with weight change we next examined 20 independent mice and assessed the relationship between bacterial levels using qPCR prior to DSS treatment and weight change observed at day 10. This analysis identified a positive correlation between weight change and pre-DSS A. faecis abundance (R2 0.3679; p = 0.0046) (Fig 3e). Similarly, a negative correlation was observed between weight change and D. muricolitica abundance (R2 0.3626; p = 0.0050) (Fig 3e). This independent confirmation provides further validation of the relationship between these species and the severity of DSS colitis; however, additional work is needed to confirm their roles as pathobionts.
Global distribution and prevalence in other mouse facilities
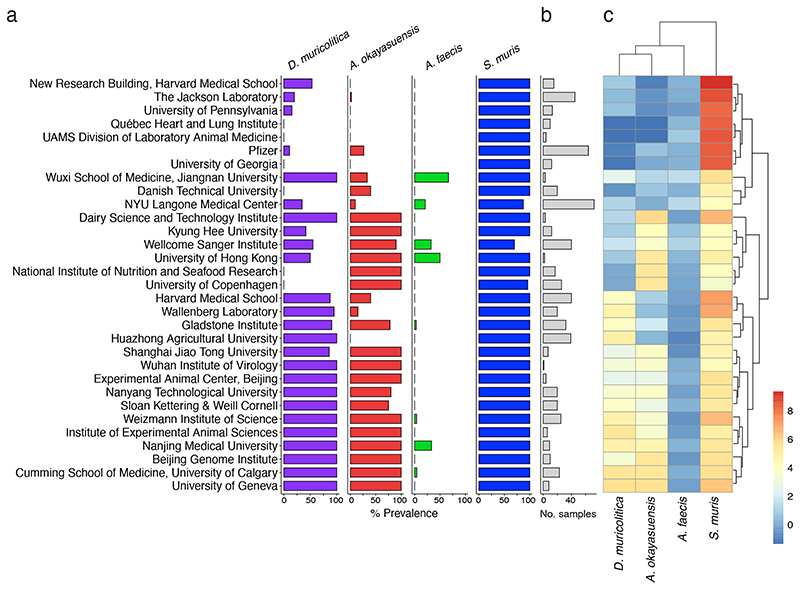
As D. muricolitica and A. okayasuensis affected the outcome of DSS colitis in our mouse facility, we next explored the presence and distribution of these bacteria in other mouse facilities around the world. To do so, we first generated a global representation of the laboratory mouse intestinal microbiomes by curating 582 publicly available shotgun metagenome samples from “control”, SPF mice. We combined this dataset with shotgun metagenomes from faeces of 40 mice at the Wellcome Sanger Institute to yield a global mouse gut microbiome dataset that covers 31 institutes across 12 countries. We then determined the prevalence and abundance of the health-associated bacteria S. muris and A. faecis and disease-associated bacteria D. muricolitica and A. okayasuensis in this mouse microbiome dataset.
D. muricolitica and A. okayasuensis were both dominant members of the mouse microbiome, present in 54.5% and 47.9% of samples with a mean abundance of 0.52% and 0.48% reads per sample respectively. Importantly, D. muricolitica and A. okayasuensis were each detected in 80.6% (25/31) of the institutes included in the analyses (Fig. 4), and there were only 3 institutes that did not contain either of these species. For comparison, of the health-associated species, S. muris was the most dominant species of the mouse gut microbiota, present in 96.5% of samples with a mean abundance of 6.2% reads, while A. faecis was a relatively minor species, present in 5.9% of samples with a mean abundance of 0.22% reads. S. muris was present in all (31/31) of the institutes included in the analyses, whereas A. faecis was detected in 25.8% (8/31) of the institutes. The abundance of each species also differed between institutes (Fig. 4). Thus, we show that mouse-derived bacteria cultured from our institute are common mouse symbionts and, importantly, D. muricolitica and A. okayasuensis are common, but not ubiquitous, in animal facilities around the world.
Figure 4. Global epidemiology and intra-institutional variation of disease- and health-associated species.
Prevalence and abundance of disease- and health-associated species in SPF mice across international mouse facilities. (a) Intra-institute prevalence of D. muricolitica (purple), A. okayasuensis (red), A. faecis (green) and S. muris (blue). A species is defined as present in a sample if ≥0.01% shotgun metagenomic reads are assigned to it. Intra-institute prevalence is calculated as the percentage of samples per institute in which a species is present. (b) Bar plot indicating number of samples per institute (grey). (c) Heatmap of intra-institute mean abundance for each species. Institute names are shared with (a). For mean abundance, data are the percentage of classified reads assigned to each species with count zeros removed using Bayesian multiplicative replacement, followed by center log-ratio transformation.
Discussion
Despite experimental variability of the DSS mouse model, which limits the interpretation and translation of data, the model is increasingly used in basic research (>6000 DSS model publications on PubMed) and by the pharmaceutical and biotechnology industries. Co-housing, heterozygote mating and other experimental procedures have been used in an attempt to normalize the microbiota within experiments and ensure direct measures of the effects of mouse genetic background on disease phenotype rather than indirect and unknown microbiota-associated effects 26 . In our approach, we leveraged this variability in DSS outcome, combined with microbiome analysis, machine learning, anaerobic culturing and germ-free mouse validation experiments to gain relevant biological insight into microbiota determinants of DSS disease. While this consideration of the microbiome did not completely eliminate the impact of multidimensionality of the DSS model, we identified and undertook experimental validation of D. muricolitica and A. okayasuensis that have a dominant effect on DSS outcome.
Interestingly, D. muricolitica and A. okayasuensis are phylogenetically distinct from each other, and neither had computationally predicted virulence factors within their genomes. While it remains unclear how these isolates contribute to DSS disease, both exhibit a potential for polysaccharide utilisation that may mediate this relationship through interaction with the mucus layer. We have also noted that weight loss and intestinal pathology do not always correlate, so they should not necessarily be used as surrogates for each other. Similarly, substantial weight loss was observed in mono-colonised mice even in the absence of severe inflammation suggesting activation of key pathways alone can have substantial impact on experimental outcome. Together these insights, raise the need for further experimental study of host interactions and the biology of these important mouse specific bacteria.
We propose that researchers should consider monitoring for D. muricolitica and A. okayasuensis in their mouse facilities, as is undertaken for other immunophenotyped-affecting microbes such as SFB 27 . In some cases, pre-colonisation of animals with these bacteria if they are not present in the facility may yield better experimental control and increase confidence in this important and widely used mouse model of human disease. Overall, our work suggests application of metagenomics techniques to report the microbiota composition in addition to the genetic and disease phenotypes being described should represent a standard minimum requirement for the DSS mouse model. We have deposited the D. muricolitis, A. okayasuensis, S. muris and A. faecis strains and genomes in public repositories to support this work.
Methods
Mouse models
Mice were maintained under either germ free or specific pathogen-free conditions at the Wellcome Sanger Institute Home Office-approved facility with all procedures carried out in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986 under Home Office approval PPL No. 80/2643. Germ free mice were maintained in positive pressure isolators (Bell) with faeces tested by culture, microscopy and PCR to ensure sterility. Consumables were autoclaved at 121°C for 15 mins prior to entering the isolators. For experimentation, cages were opened in a vaporised hydrogen peroxide (Bioquell) sterilised Class II cabinet with faecal transplant and single colonised gnotobiotic lines generated by weekly oral gavage over a 3-week period with material prepared in D-PBS at 100mg/ml immediately prior to administration under anaerobic conditions (10% H, 10% CO2, 80% N) in a Whitley DG250 workstation at 37 °C 8 . Mice were maintained in sterile ISOcages (Tecniplast) and housed on ISOrack for the period of the experiment.
DSS colitis challenge
DSS was administered at a concentration of 1.5% (w/v) DSS (Affymetrix, Inc., molecular weight: average 44kDa) to drinking water for 7 days, followed by 3 days with regular drinking water to C57BL6/6N mice aged between 7 and 16 weeks (297 male, 282 female; median 11 weeks; SD 12 days). Mice were weighed every day and culled if weight loss reached 20% of starting weight. Average weight loss curves were censored for each group when the first member of the group reached this end-point to avoid misleading calculations of the average using the remaining members. No statistical methods were used to pre-determine sample sizes in these experiments.
Histological assessment of intestinal inflammation
DSS-treated SPF mice were sacrificed at day 10 by cervical dislocation, and samples from mid- and distal colon taken. Tissue sections were fixed in buffered 10% formalin; paraffin-embedded; cut; and stained with haematoxylin and eosin. Colon histopathology was blind-graded semi-quantitatively on a scale from zero to three, for four criteria: (1) degree of epithelial hyperplasia/damage and goblet cell depletion, (2) leukocyte infiltration in lamina propria, (3) area of tissue affected, and (4) presence of markers of severe inflammation, including crypt abscesses, submucosal inflammation, and oedema. Scores for individual criteria were added for an overall inflammation score of between zero and twelve for each sample. Scores from mid and distal colon were then averaged to obtain inflammation scores for each mouse colon.
For monocolonised germ-free mice, animals were euthanized 7 days after DSS administration, and samples were taken from the caecum and mid colon. Tissue sections were fixed in methacarn, paraffin-embedded, sectioned, and stained with haematoxylin and eosin. Colon and caecum histopathology was graded on a semi-quantitative scale by a pathologist blinded to the groups. The criteria scored were 1) degree of epithelial hyperplasia/damage and goblet cell depletion, 2) severity of leukocyte infiltration in lamina propria 3) extent of the inflammation, and 4) presence of markers of severe inflammation, including crypt abscesses, submucosal inflammation and oedema. Each criterion was scored on a scale of zero to three based on previously described thresholds 28 and the sum of the individual scores was recorded as an indication of the overall inflammation. For all animal experiments, treatments were randomised by cage by researchers blinded to treatment conditions. For statistical analysis, distribution was assumed to be normal but this was not formally tested
Bacterial Culturing
Faecal samples and bacterial isolates were cultured for anaerobic gastrointestinal bacteria under anaerobic conditions (10% H, 10% CO2, 80% N) in a Whitley DG250 workstation at 37 °C 8 . Faecal samples were homogenized in reduced PBS (0.1 g per ml PBS), serially diluted and plated directly onto YCFA agar supplemented with 0.002 g ml−1 each of glucose, maltose and cellobiose. Colonies were picked, restreaked to purity and identified using 16S rRNA gene sequencing.
Microbiota profiling and analysis
Faecal samples were collected directly from mice, and immediately stored at -80°C until DNA extraction. DNA was extracted from faecal samples using FastDNA Spin Kit for Soil (MPBio) and stored at -20 °C until metagenomic sequencing. DNA samples were quantified using a Qubit 4 Fluorometer (Thermo Fisher). 16S rRNA amplicon based profiling was performed using the FastDNA Spin Kit for Soil (MP Biomedicals) on 300mg of faecal sample from each mouse. The V1-V2 region of the 16S rRNA genes was amplified with Q5 High-Fidelity Polymerase Kit (New England Biolabs). (F’:AATGATACGGCGACCACCGAGATCTACAC-TATGGTAATT-CC-AGMGTTYGATYMTGGCTCAG; R’:CAAGCAGAAGACGGCATACGAGATACGAGACTGATTAGTCAGTCAGAAGCTGCCTCCCGTAGGAG). Each PCR amplification was performed as four independent amplifications and pooled in equimolar amounts for 150bp paired end sequencing with the Illumina MiSeq platform. For shotgun metagenomic sequencing, samples with >100 ng DNA material proceeded to paired-end (2 × 150 bp) metagenomics sequencing on the HiSeq 4000 platform as per standard manufacturer library preparation and sequencing protocols.
Metagenomic Data Analysis
16S rRNA amplicon sequences were analysed with mothur MiSeq SOP v.1.42.3 using SILVA v.132 29 . The 16S rRNA gene alignments were used to determine a maximum likelihood phylogeny using FastTree v.2.1.10 30 . Phylogenetic trees were visualized and edited using iTOL 31 . Shotgun metagenomic reads were filtered for quality and adaptor sequences using KneadData v0.7.3 with default settings. Host reads were removed from samples using the GRCm39 reference genome and Bowtie2 v2.3.5. In addition, reads were aligned to the Phi X 174 genome and removed. Taxonomic classification of metagenomic reads was performed using Kraken2 v2.0.8 and a custom database built from the genomes of the Mouse Gut Bacterial Catalogue.
All data analyses were performed in R v4.0.2. Species abundant at ≥0.01% reads were considered present in a sample and OTUs were considered statistically different at p < 0.05. For shotgun metagenomic sequencing, relative abundance of species was corrected using Bracken v2.5.2. Bar plots for species prevalence data and sample numbers were generated using the ggplot2 v3.3.3 package. To quantify and visualise intra-institute abundance, species abundance data for each sample underwent Bayesian multiplicative replacement of count zeros using the zCompositions v1.3.4 package, and the data transformed using centre-log ratio normalisation. Mean abundance per sample was then calculated and visualised using the pheatmap v1.0.12 package. Principal component analyses were performed in R and Linear regression analysis was performed using the Linear Regression function in python scikit-learn 0.20.4. Alpha diversity was assessed using Vegan 2.5.6. 30 . Correlation analysis was performed using Spearman’s rank correlation coefficient.
Curation of public metagenome samples
To assess the abundance of the health- and disease-associated bacteria in mice from different institutes, we curated publicly available faecal shotgun metagenomes for healthy, “control”, SPF mice from NCBI and the ENA. These samples included mice from different genetic backgrounds and strains, but samples were excluded if mice were younger than 3 weeks, had received antimicrobial treatment or dietary intervention, or had a gene knockout. In total 582 faecal shotgun metagenomes were curated.
Quantitative PCR Quantification of bacterial load
Anaerostipes faecis and Duncaniella muricolitica were quantified in the faeces of SPF mice using quantitative PCR with custom primers. Primers were designed using the taxon-linked gene catalogue of the Mouse Gastrointestinal Bacterial Catalogue (MGBC) 32 . Briefly, we identified gene targets that were both unique to and highly conserved in genomes of each species. To ensure target specificity, we selected gene clusters that shared <50% sequence identity with all non-target species sequences. Gene targets were defined as highly conserved if they were identified in 100% of high-quality target species genomes in the MGBC. Furthermore, only single copy number gene targets were considered. Sequences for selected gene products were then clustered at 90% sequence identity and used as templates for designing PCR primers with NCBI Primer-BLAST 33 , ensuring specificity against the NCBI non-redundant gene catalogue. Two primers for each species were used in conjunction for qPCR.
Primers: A14_1 (gene target: MGBC000029_00009), forward 5’-AGCTGTTCAGGTGTCTGATCTT-3’, reverse 5’-CGGTCCACTTCGCAGTATCA-3’, product size 74 bp; A14_2 (gene target: MGBC000029_00377), forward 5’-ATGCAGCACTGGGAGATTCA-3’, reverse 5’-CTGCGTGTCCACAGAAGAGT-3’, product size 70 bp; A60_1 (gene target: MGBC109741_00862), forward 5’-CCATCTTTTCGGACGGTGGA-3’, reverse 5’-CGAGGTCTCCCTGAAACGAC-3’, product size 125 bp; A60_2 (gene target: MGBC140600_00802), forward 5’-AAAACGGTGACAGCTCGGAA-3’, reverse 5’-GTCTCTTTTTGGCGGATGGG-3’, product size 93 bp.
DNA was extracted from faeces using the ‘FastDNA Spin Kit for Soil’ (MPBio) according to manufacturer’s instructions, and DNA eluted into 100uL ddH2O. Eluted DNA was then diluted 1:50 and qPCR performed using SYBR Green chemistry (Thermo Fisher). To approximate the relative abundance of target species in SPF faeces, CT values were normalised to those of a universal bacterial 16S primer (F: 5’-GTGSTGCAYGGYTGTCGTCA-3’; R: 5’-ACGTCRTCCMCACCTTCCTC-3’).
Intestinal immune cell isolation
Large intestines were excised and separated from mesentery and fat. Transluminal sections were taken from the caecum and distal colon for histology. The intestines were opened longitudinally, cleared of faeces, and then washed three times in cold PBS. To isolate the intraepithelial immune populations, the large intestines were cut into 1cm pieces, washed in cold PBS, and then incubated in 10 mL of PBS + 1 mM dithiothreitol for 10 minutes. Tissues were manually disrupted via shaking and then strained, collecting the supernatant. Tissues were then incubated in 10 ml of PBS + 30 mM EDTA + 10 mL HEPES at 37°C at 200 rpm, before being shaken and strained. The supernatants from these two steps were pooled, filtered at 70 μm and the filtrate fractionated using a discontinuous Percoll gradient (80%/40%). Epithelial cells were isolated from the surface of the Percoll, and the intraepithelial immune cells isolated from the interface.
To access the lamina propria compartment, the tissues were manually chopped and digested in HBSS + 25 mM HEPES + 1 mM sodium pyruvate containing 0.05 mg/mL Collagenase VIII (Sigma) and 50 μg/ml DNase I (Sigma) for 1 hour at 37°C at 80 rpm. Samples were mechanically disrupted and filtered at 70 μm. The filtrate was fractionated using a discontinuous Percoll gradient (80%/40%). Lamina propria immune cells were isolated from the interface.
Flow cytometry
Intestinal and mesenteric lymph node immune cell populations were characterised by flow cytometry. For intracellular cytokine staining, cells were incubated in T cell media (RPMI, 10% FBS, 1 mM sodium pyruvate, 2mM GLUTamax, 1X non-essential amino acids, 0.1 mM 2-β-mercaptoethanol, 10 mM HEPES, 1%Penicillin/Streptomycin) + 1X Cell Stimulation Cocktail with Protein Inhibitors (eBioscience) for 3 hours at 37°C before staining. Non-viable cells were stained using Live/Dead Fixable Aqua. Cells were permeabilised with CytoFix/CytoPerm (BD) followed by intracellular staining in PermBuffer (eBioscience).
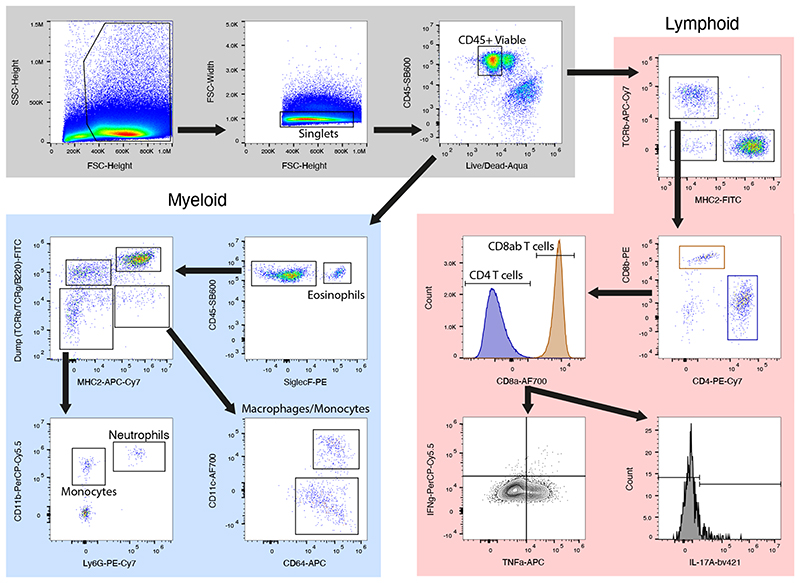
All antibodies were purchased from eBioscience unless otherwise indicated. Antibodies used were CD45-SB600, TCRb-APC-Cy7, MHCII-FITC, CD4-PE-Cy7, CD8a-AF700, CD8b-PE, IFNg-PerCP-Cy5.5, TNFa-APC, IL-17A-bv421 (BioLegend), CD3e-FITC, TCRg-FITC, B220-FITC, MHCII-APC-e780, CD11b-PerCP-Cy5.5, CD11c-AF700, CD64-APC (BioLegend), SiglecF-PE (BD), Ly6G-Pe-Cy7 (BD) and F4/80-e450. Sample data were acquired with an Attune NxT flow cytometer coupled with an Attune CytKick Max autosampler. Data were analysed using FlowJo v10. Antibody details and concentrations are included in Supplementary Table 1.
Bacterial species characterisations
While genomes are publicly available for the species described in this paper and they are represented by genomic species clusters in the Genome Taxonomy Database (GTDB) (A14=Anaerostipes sp000508985 (GCF_000508985.1: 98.76% ANI), A43=CAG-485 sp002362485 (GCF_003833075.1: 98.66% ANI), A60=Duncaniella sp001689575 (GCF_003762875.1: 96.19% ANI), A61=Alistipes sp002362235 (GCA_002362235.1: 98.54% ANI)), these species have not yet been characterised nor have these names been validly published according to the International Code of Nomenclature of Prokaryotes. As these species are uncharacterised they are referred to as previously undescribed in this manuscript. Phenotypic data for the capacity of each species to utilise 95 substrates as sole carbon sources were attained using the BioLog platform (see ‘BioLog analysis’ section for methodology). Isolate genomes were functionally annotated using eggNOG-mapper v2 and InterProScan v5. We describe these species and propose new names below.
Description of “Alistipes okayasuensis” sp. nov
Alistipes okayasuensis (o’ka.ya.su.en.sis. N.L. masc./fem. adj. okayasuensis, named after Isao Okayasu, who first described dextran sulphate sodium colitis as a model for ulcerative colitis in mice. Phylogenomic analyses place strain A61T in the Alistipes cluster. The dDDH value between Alistipes onderdonkii and Alistipes putredinis is 24.4%, while the dDDH values between strain A61T, Alistipes onderdonkii and Alistipes putredinis are 25.2% and 21.4% respectively. The G+C content difference between the genome of strain A61T and Alistipes onderdonkii, its closest phylogenomic neighbour, is 1.39%, while their 16S rRNA gene sequence identity is 96.03%. These analyses strongly indicate that “Alistipes okayasuensis” is a separate species within the genus Alistipes.
This strain is a strict anaerobe. Carbon source utilisation analysis was combined with reconstruction of KEGG metabolic pathways (55.58% of 2022 predicted ORFs could be annotated) to functionally characterise the strain. The strain encodes 47 genes predicted to be carbohydrate-active enzymes (CAZymes), including 29 glycoside hydrolases: seven beta-glucosidases (GH3, EC 3.2.1.21); five beta-galactosidases (EC 3.2.1.23); five endohydrolytic alpha-glucosidases (GH13) including four alpha-amylases (EC 3.2.1.1); three exohydrolytic alpha-glucosidases (GH31, EC 3.2.1.20); three cellulases (EC 3.2.1.4); three mannan endo-1,4-beta-mannosidase (EC 3.2.1.78); and an alpha-L-fucosidase (EC 3.2.1.51). Despite this predicted hydrolytic potential, the strain shows limited capacity to use disaccharides and oligosaccharides as sole carbon sources. While the strain tested positive for utilising alpha-D-lactose, sucrose and maltotriose, it tested negative for utilisation of D-cellobiose, dextrin, cyclodextrin, gentiobiose, lactulose, maltose, D-melezitose, D-melibiose, palatinose, D-raffinose, stachyose, D-trehalose, fucose or turanose.
The strain is predicted to encode enzymes for metabolism of L-histidine to L-glutamate via 4-Imidazolone-5-phsophate (EC 4.3.1.3, EC 4.2.1.49, EC 3.5.2.7, EC 2.1.2.5). In support of these predictions, the strain is capable of utilising urocanate as a sole carbon source. In-keeping with other members of the Alistipes genus, this strain is predicted to encode a putative tryptophanase (EC 4.1.99.1) that would allow it to hydrolyse tryptophan to indole. The strain is also predicted to encode a butyrate phosphotransferase (EC 2.3.1.19) and a butyrate kinase (EC 2.7.2.7), indicating potential to metabolise butyrate. The genome is 2,329,524 bp with a G+C content of 56.45 mol%. We propose the name “Alistipes okayasuensis” for this previously undescribed species, in reference to the origins of the DSS colitis model and reflecting this species’ association with poor outcomes of this disease model. The type strain is A61T (=CCUG 75087T = DSM 112987T).
Description of “Anaerostipes faecis” sp. nov
Anaerostipes faecis (fae’cis. L. gen. fem. n. faecis, of faeces, referring to faecal origin). The closest phylogenomic neighbour of strain A14T is Anaerostipes caccae with a dDDH value of 48.40%. The pairwise 16S rRNA gene sequence identity of A14T and A. caccae DSM 14662T is 98.16%. These analyses indicate that A14T is a separate species within the genus Anaerostipes. The genome is 3,200,246 bp with a G+C content of 43.77 mol%. We propose the name “Anaerostipes faecis” for this new species. The type strain is A14T (=CCUG 75084T).
The strain is strictly anaerobic. Reconstruction of KEGG metabolic pathways (55.4% of 3222 predicted ORFs could be annotated) indicates that the strain can produce butyrate via a butyrate phosphotransferase/butyrate kinase operon (EC 2.7.2.7, EC 2.3.1.91), in keeping with the genus type species A. caccae. Likewise, the strain is predicted to encode an arginine dihydrolase (EC 3.5.3.6). In contrast to the genus description however, this strain is not predicted to encode an alpha-galactosidase (EC 3.2.1.22) nor a phosphoamidase (EC 3.9.1.1).
The strain shows limited capacity to utilise monosaccharides as sole carbon sources: it had a robust phenotype for utilising 3-methyl-D-glucose, but tested negative for its ability to utilise other monosaccharides including D-fructose, D-galactose, D-mannose or D-glucose as sole carbon sources. Notably, the strain is predicted to encode the complete Leloir pathway for galactose metabolism as well as multiple beta-galactosidases (EC 3.2.1.23), indicating that the strain might lack a galactose transporter but could metabolise galactose in different contexts such as downstream of other pathways of carbohydrate metabolism. The strain can utilise some disaccharides, testing positive for sucrose utilisation, potentially via multiple oligo-1,6-glucosidases (EC 3.2.1.10), D-trehalose utilisation, potentially via three trehalases (EC 3.2.1.28, GH37, GH65), and turanose utilisation. However, the strain tested negative for its ability to utilise alpha-D-lactose, lactulose, maltose, D-melibiose, gentiobiose, D-cellobiose or palatinose. Additionally, the strain tested negative for utilisation of the trisccharides maltotriose, D-melezitose, D-raffinose, and the tetrasaccharide stachyose – potentially due to the absence of a predicted alpha-galactosidase. The strain could utilise beta-cyclodextrin, but tested negative for utilisation of dextrin and alpha-cyclodextrin. The strain is capable of utilising alpha-ketovaleric acid and D-malic acid, as well as certain amino acids and derivatives including L-threonine, L-asparagine, L-serine and the dipeptides glycyl-L-proline and L-alanyl-Lthreonine. The strain tested negative for utilisation of L-glutamine, L-glutamate, L-valine and L-alanine.
Description of “Duncaniella muricolitica” sp. nov
Duncaniella muricolitica (mu.ri.co’li.ti.ca. L. gen. n. muris, of the mouse; Gr. n. kolon, colon; Gr. suff. -tikos -ê -on, suffix used with the sense of pertaining to; N.L. fem. adj. muricolitica, pertaining to the colon of the mouse). The closest phylogenetic neighbour according to pairwise 16S rRNA gene analyses is Duncaniella dubosii with a sequence identity of 94.04%. The 16S gene sequence identity values between D. dubosii H5T and other species of the genus, D. muris DSM 103720T and D. freteri TLL-A3T, are 95.14% and 93.00% respectively, indicating that A60T is a separate species within the Duncaniella cluster. The dDDH value between A60T and D. dubosii H5T is 28.10%, confirming a separate species status.
Recent work benchmarking an average nucleotide identity 1 approach to define genus boundaries found that the genus inflection points of members of the order Bacteroidales had relatively low values for alignment fraction (AF) and ANI (AF:ANI: Bacteroides fragilis=0.215:71.23; Porphyromonas asaccharolytica=0.105:68.12; Prevotella melaninogenica=0.195:72.07). The corresponding values between A60T and the genus type species D. muris are 0.243:80.85, providing further evidence that A60T belongs to the genus Duncaniella.
The strain is an obligate anaerobe. Reconstruction of KEGG metabolic pathways (47.2% of 3461 predicted ORFs could be annotated) indicated that strain is enriched for enzymes with glycoside hydrolase activity, encoding 47 in total. The strain is able to utilise dextrin, beta-cyclodextrin and multiple oligosaccharides including D-melezitose, maltotriose, amygdalin, maltose, gentiobiose (EC: 3.2.1.21), turanose, and D-cellobiose (EC: 2.4.1.20; EC: 3.2.1.21). The strain is capable of utilising D-raffinose, and is predicted from its genome to produce sucrose as an intermediate (EC: 3.2.1.22, EC: 3.2.1.20) instead of stachyose or melibiose. Additionally, the strain tested negative for utilisation of both stachyose and melibiose. In our study, the strain tested negative for an ability to utilise lactose as a single carbon source, even though it encodes seven predicted beta-galactosidases (EC 3.2.1.23). The strain is predicted to encode 36 glycosyltransferase enzymes. Among these are seven GT2 family enzymes including two dolichyl-phosphate beta-D-mannosyltransferases (EC: 2.4.1.82).
The strain can utilise a variety of monosaccharides as single carbon sources, including alpha-D-glucose, alpha-D-glucose 1-phosphate, 3-methyl-D-glucose, D-galactose, alpha-methyl-D-galactose, D-mannose, adonitol, arabinose, D-gluconate, D-arabitol, and D-sorbitol. It tested negative for an ability to utilise fucose, alpha-D-lactose, D-fructose, lactate, lactulose, D-trehalose or L-rhamnose as sole carbon sources. The strain is predicted to encode the pathway for metabolism of L-histidine to L-glutamate via 4-Imidazolone-5-phsophate (EC 4.3.1.3, EC 4.2.1.49, EC 3.5.2.7, EC 2.1.2.5), and tests positive for utilisation of urocanate. The genome is 4,055,027 bp with a G+C content of 50.9 mol%. Following functional and genomic characterisation of our isolate, we propose the name “Duncaniella muricolitica” for this species, in reference to its association with poor prognosis in the murine DSS model of colitis. The type strain is A60T (=CCUG 75086T = DSM 112986T).
Description of “Sangeribacter” gen. nov
Sangeribacter (San.ge.ri.bac’ter. N.L. gen. masc. n. Sangeri, of Sanger; N.L. masc. n. bacter, rod; N.L. masc. n. Sangeribacter, a rod-shaped bacterium named after Frederick Sanger (1918-2013) and the institute where this genus was first described). 16S rRNA gene sequence analyses place this strain in the Muribaculaceae family, and “Sangeribacter” possess all features of this family. The pairwise sequence identity of the 16S rRNA gene of A43T with 16S genes from Paramuribaculum intestinale DSM 100749T, Muribaculum intestinale YL27T and Duncaniella muris DSM 103720T is 88.90%, 88.22% and 87.39% respectively. The dDDH value between A43T and Paramuribaculum intestinale DSM 100749T is 35.5% and the difference in G+C mol% is 6.37%. Together these analyses strongly indicate that “Sangeribacter” is a separate genus. The type species, Sangeribacter muris, is one of the most dominant species in the mouse gut microbiota, representing up to 60% of classified reads in some mouse faecal shotgun metagenome samples. It is also highly prevalent, present in 77% of 1,926 samples.
Description of “Sangeribacter muris” sp. nov
Sangeribacter muris (mu’ris. L. gen. masc./fem. n. muris, of the mouse, the species was first isolated from a mouse). The strain is a strict anaerobe. It is predicted to encode 66 CAZymes including 42 glycoside hydrolases. The strain tests positive for utilisation of a wide repertoire of glycans and carbohydrates, including dextrin (EC 3.2.1.3), alpha- and beta-cyclodextrin, stachyose (EC 3.2.1.22), D-raffinose (EC 3.2.1.26), maltotriose, D-melezitose, amygdalin (EC 3.2.1.22), D-cellobiose (EC 3.2.1.21, EC 2.4.1.20), gentiobiose (EC 3.2.1.21), D-melibiose, palatinose, D-trehalose (EC 2.7.1.201, EC 2.4.1.64, EC 3.1.3.12), turanose, sucrose (EC 2.7.1.211), D-glucosaminate, lactulose, alpha-D-lactose, alpha-D-glucose, alpha-D-glucose 1-phosphate, 3-methyl-D-glucose, D-glucosaminic acid, alpha- and beta-methyl-D-glucoside, D-fructose (EC 2.7.1.4), D-mannitol (EC 1.1.1.67), D-mannose, D-galactose, alpha- and beta-methyl-D-galactoside, D-galacturonic acid, L-sorbitol (EC 1.1.1.14), and L-fucose (EC 3.2.1.51). This strain tested negative for its ability to utilise adonitol, dulcitol or D-gluconate as single carbon sources.
The strain tested positive for being able to utilise certain amino acids and their derivatives, including L-phenylalanine, L-asparagine, L-glutamate, L-glutamine, L-alanine, and L-valine, as well as dipeptides including L-alanyl-L-threonine, L-alanyl-L-glutamine, L-alanyl-L-histidine, glycyl-L-aspartate, glycyl-L-glutamine and glycyl-L-proline. It tested negative for an ability to utilise glycyl-L-proline, L-serine, L-aspartate or L-threonine as single carbon sources. The strain is predicted to encode a tryptophanase (EC 4.1.99.1) indicating that it has potential to hydrolyse tryptophan to indole. The strain tested negative for utilisation of propionate or hydroxybutyrate as sole carbon sources, indicating that it may not consume these short chain fatty acids in vivo. The genome is 3,532,505 bp with a G+C content of 46.70 mol%. We propose the name “Sangeribacter muris” for this new species, reflecting its high abundance and prevalence in mice and its initial isolation from a murine host. The type strain is A43T (=CCUG 75085T)
BioLog analysis
Isolates were streaked on YCFA agar media and grown overnight. Cotton swabs were used to remove colonies which were then inoculated in AN-IF Inoculating Fluid (Technopath product code 72007) to a turbidity of 65% using a turbidimeter. Then 100ul was pipetted into each well of Anaerobe AN Microplates (Technopath, product code 1007) which contain 95 different carbon sources. The plates were sealed in PM Gas Bags (Technopath, product number 3032) and run on the Omnilog system for 24 hours. For each isolate, 3-5 replicates were run on different days from different starting colonies. Data were analysed using the CarboLogR application 34 . The quality filtered CarboLogR output is provided in Supplementary Table 2.
Extended Data
Extended Data Fig. 1. Lineage specific variation across C57BL/6 mice.
Correlation between variation in weight change and average histology score across wild-type mice (n=579) with (a) age in weeks, (b) mouse weight at experiment commencement and (c) lineage. Boxplot showing minimum, 25th percentile, median, 75th percentile and maxima.
Extended Data Fig. 2. Phyla level microbiome composition Pre- and Post-DSS.
Individual sample relative abundance of Bacteroides (red), Firmicutes (green) and Proteobacteria (blue) and other (grey) in (a) pre- and (b) post-DSS samples by 16S rRNA amplicon sequencing.
Extended Data Fig. 3. Family level microbiome composition Pre- and Post-DSS.
Individual sample relative abundance of dominant families identified through 16S rRNA amplicon sequencing. Data for pre-DSS (n=77) and post-DSS (n=77) samples is shown.
Extended Data Fig. 4. Genomic Characterisation of Bacterial Isolates.
Phylogenetic tree showing relationship of A. faecis (blue circle), S. muris (blue triangle), D. muricolitica (red circle) and A. okayasuensis (red triangle) with known type strains (yellow star).
Extended Data Fig. 5. Mono-colonisation of germ-free mice.
(a) Bacterial colonisation at day 0 prior to DSS treatment in mice pre-colonised with either A. okayasuensis (red), A faecis (green), D. muricolitica (purple) or S. muris (blue) and germ-free controls (PBS; black) presented as CFU/g of faecal material. No bacterial colonies were detected 1 in the PBS control mice. Flow cytometric analysis of (b) monocytes/macrophages (B220/CD3e/TCRg-MHC2+CD64+CD11c+) and (c) other immune cell populations from the large intestine lamina propria of control germ-free mice (PBS) or mice mono-colonised with A. faecis or D. muricolitica without DSS challenge.
Extended Data Fig. 6. Flow cytometric gating strategy.
Gating strategy for flow cytometric quantification of myeloid (blue) and lymphoid (red) immune cell populations in the large intestine lamina propria. IFN, interferon; TNF, tumour necrosis factor; IL, interleukin.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust [098051] and the Australian National Health and Medical Research Council [1091097, 1159239 and 1141564 to SF]. VP is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society [206245/Z/17/Z]. BBJ is supported by a studentship from the Rosetrees Trust [A2194]. KJM is supported by a Wellcome Trust Investigator Award (102972/Z/13/Z) We are grateful to the Wellcome Sanger Institute Pathogen informatics and Research Support Facility for supporting this research.
Funding for open access charge
Wellcome Sanger Institute.
Footnotes
Author Contributions
SC, TL, SF, VP conceived the study and study design. KH, SC, GN, CB, BBJ and AS performed animal experiments. SF, BBJ, and NK undertook genomic and computational analyses. SF, GN, MS and AA cultured bacteria, HH performed histological analyses, BM and KM designed and executed the DSS screen. BBJ, AS and VP performed mouse immune cell isolations and profiling. GD, DA and TL established the study. All authors contributed to preparing the manuscript.
Competing Interest Statement
TDL is a founder and CSO of Microbiotica. SCF is an advisor to Biomebank. The other authors declare no competing financial interests.
Data availability
Metagenomic sequencing data is deposited in the ENA under project PRJEB50449. Key strains were deposited in the University of Gothenburg Culture Collection (CCUG) or the DSMZ German Collection of Microorganisms (DSM) under the following identifiers Alistipes okayasuensis (CCUG 75087T|DSM 112987T), Duncaniella muricolitica (CCUG 75086T|DSM 112986T), Sangeribacter muris (CCUG 75085T) and Anaerostipes faecis (CCUG 75084T).
Code availability statement
No custom code was used in this research.
References
- 1.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCafferty J, et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen HB, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 5.Almeida A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Jumpstart Reference Strains, C et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne HP, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forster SC, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou Y, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37:179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagkouvardos I, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1:16131. doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, et al. The Mouse Gut Microbial Biobank expands the coverage of cultured bacteria. Nat Commun. 2020;11:79. doi: 10.1038/s41467-019-13836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy U, et al. Distinct Microbial Communities Trigger Colitis Development upon Intestinal Barrier Damage via Innate or Adaptive Immune Cells. Cell Rep. 2017;21:994–1008. doi: 10.1016/j.celrep.2017.09.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullberg MC, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12-and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/IAI.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/-mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Chirlaque C, et al. Germ-free and Antibiotic-treated Mice are Highly Susceptible to Epithelial Injury in DSS Colitis. J Crohns Colitis. 2016;10:1324–1335. doi: 10.1093/ecco-jcc/jjw096. [DOI] [PubMed] [Google Scholar]
- 19.Llewellyn SR, et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology. 2018;154:1037–1046.:e1032. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surana NK, Kasper DL. Moving beyond microbiome-wide associations to causal microbe identification. Nature. 2017;552:244–247. doi: 10.1038/nature25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015;8:476–486. doi: 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Neville BA, Forster SC, Lawley TD. Commensal Koch’s postulates: establishing causation in human microbiota research. Curr Opin Microbiol. 2018;42:47–52. doi: 10.1016/j.mib.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold IC, et al. CD11c(+) monocyte/macrophages promote chronic Helicobacter hepaticus-induced intestinal inflammation through the production of IL-23. Mucosal Immunol. 2016;9:352–363. doi: 10.1038/mi.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson SJ, et al. Comparison of Co-housing and Littermate Methods for Microbiota Standardization in Mouse Models. Cell Rep. 2019;27:1910–1919.:e1912. doi: 10.1016/j.celrep.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 27.Farkas AM, et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods. 2015;421:104–111. doi: 10.1016/j.jim.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erben U, et al. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- 29.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beresford-Jones BS, et al. Functional mapping enables translation between the mouse and human gut microbiotas. Cell host and microbe Accepted. 2021 doi: 10.1016/j.chom.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye J, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vervier K, Browne HP, Lawley TD. CarboLogR: a Shiny/R application for statistical analysis of bacterial utilisation of carbon sources. bioRxiv. 2019:695–676. doi: 10.1101/695676. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic sequencing data is deposited in the ENA under project PRJEB50449. Key strains were deposited in the University of Gothenburg Culture Collection (CCUG) or the DSMZ German Collection of Microorganisms (DSM) under the following identifiers Alistipes okayasuensis (CCUG 75087T|DSM 112987T), Duncaniella muricolitica (CCUG 75086T|DSM 112986T), Sangeribacter muris (CCUG 75085T) and Anaerostipes faecis (CCUG 75084T).
No custom code was used in this research.