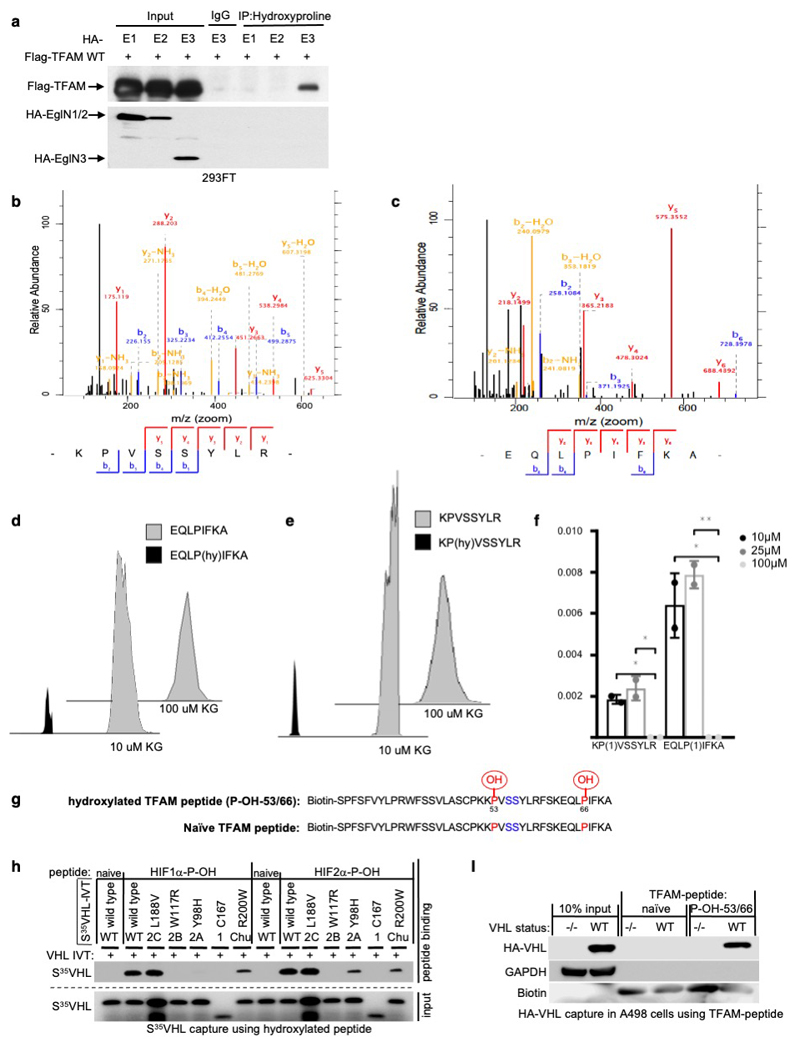
Extended Data Fig. 4. TFAM is hydroxylated by EglN3 at Proline 53/66 causing pVHL recognition.
(a) Immunoprecipitation using antihydroxyproline antibody (HydroxyP) from 293FT cells that were transiently transfected with plasmids encoding Flag-TFAM and HA-EGLN1, HA-EGLN2 and HA-EGLN3. Immunoblots show co-immunoprecipitation of Flag-TFAM. n = 3 biological independent experiments. (b-c) Mass spectrometry of unmodified biotinylated TFAM-peptide-30-70. Shown is the representative fragmentation peptide spectra of non-hydroxylated Biotin-KPVSSYLR (b) and non-hydroxylated Biotin-EQLPIFKA (c). (d,e) Extracted ion chromatogram of biotinylated unmodified and mono hydroxylated proline residues 53 (d) or proline residues 66 (e) TFAM peptide following an in vitro hydroxylation reaction with EglN3 with indicated concentration of ϒ-ketoglutarate (KG). Control indicates unmodified biotinylated TFAM-peptide that was not subjected to EGLN3 hydroxylation. (f) Hydroxylation levels of proline residues 53 and 66 of TFAM peptide following hydroxylation with EGLN3 generated via IVT with indicated concentration of KG. Data are presented as mean values ± SD. n = 3 biological experiments. One way ANOVA Tukey's Multiple Comparison Test. *p <0.05, **p <0.01. p=0.0288, p=0.0143, p=0.0148, p=0.0082. (g) Schematic illustration of synthetic biotinylated TFAM peptide hydroxylated at P-OH-53 and P-OH-66 and naïve TFAM peptide. (h) Autoradiograms showing recovery of 35S-labeled VHL protein (WT) or corresponding disease mutants (as indicated) bound to biotinylated HIF1α peptide (residues 556 to 575) with hydroxylated proline 564 (HIF1α-P-OH) and HIF2α peptide (residues 521 to 543) with hydroxylated proline 531 (HIF2α-P-OH). Biotinylated HIF1α and HIF2α naïve peptides were used as negative controls. n = 3 biological independent experiments. (i) Peptide pulldown using biotinylated TFAM-P-OH-53/66 peptide incubated with whole-cell lysates from A498 cells expressing HA-VHL WT or empty control. Biotinylated TFAM naïve peptide was used as negative control. n = 3 biological independent experiments.