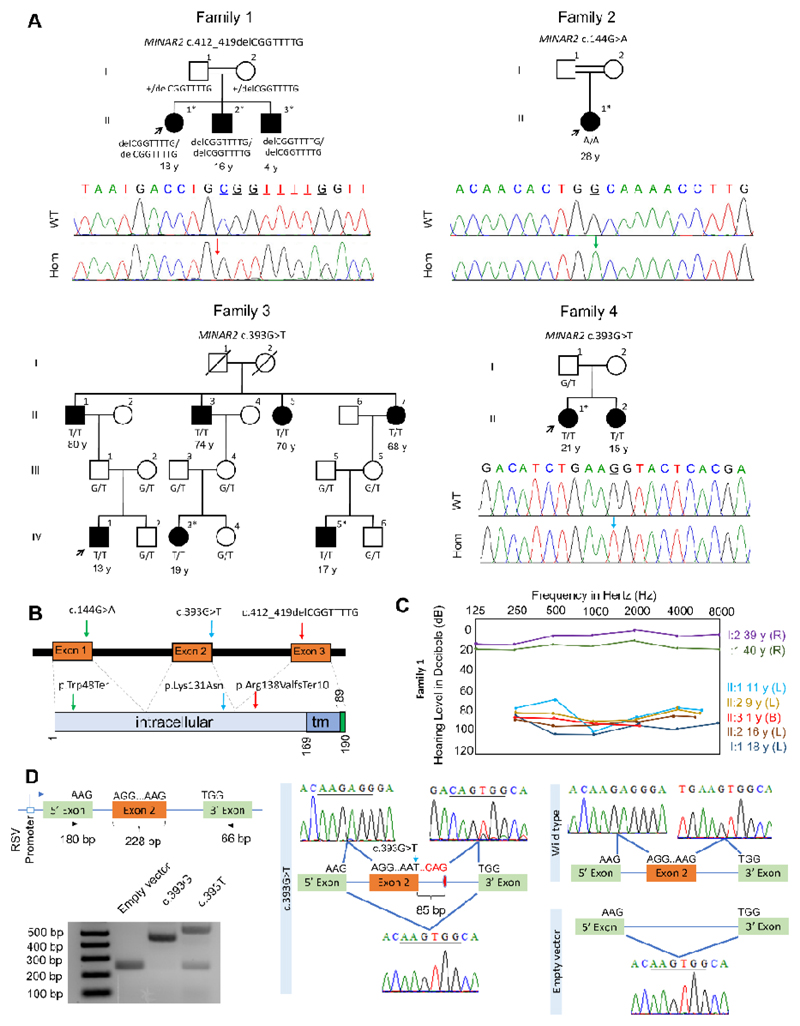
Figure 1. Families, MINAR2 variants, and the effects of the c.393G>T variant on splicing.
(A) Pedigrees and segregation of the MINAR2 variants in families. Filled symbols denote affected individuals and double lines indicate first cousin consanguinity. Electropherograms show the identified variant. The wildtype traces are from an unrelated individual. WT: Wildtype, Hom: Homozygous mutant; exome/genome sequencing was performed in individuals marked with an (*) asterisk. (B) Locations of the identified variants. tm: transmembrane (C) Audiogram of the Family 1 (R: Right ear, L: Left ear, B: Bilateral). (D) MINAR2 exon 2 inserted into a vector consisting of 5’ and 3’ exons in the exon trap assay is shown. There are larger and smaller PCR products in the c.393G>T sample compared to wildtype. Sanger sequencing confirms insertion of 85 bp at the donor site of exon 2 and skipping of exon 2.