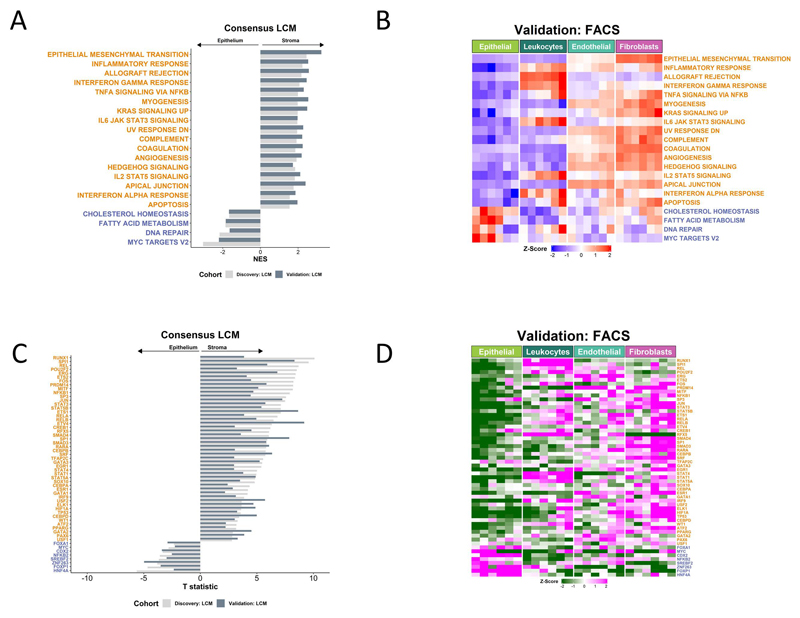
Figure 2. Stromal influence on widely used transcriptional signatures.
A Gene set enrichment analysis (GSEA) of Hallmark gene sets in LCM discovery and validation cohorts. Only gene sets significantly and concordantly enriched in stroma or epithelium in both the discovery and validation cohorts are shown (adjusted p-value < 0.02). B Heatmap of single sample GSEA (ssGSEA) scores for the Hallmark gene sets in the FACS validation cohort samples. Only the gene sets significantly and concordantly enriched in stroma or epithelium in both the LCM discovery and validation cohorts are shown (adjusted p-value < 0.02). C Transcription factors whose activity was significantly and concordantly enriched in stroma or epithelium in both the LCM discovery and validation cohorts (p < 0.05). D Heatmap of the inferred activity scores for the same transcription factors in the FACS validation cohort. For all panels in Figure 2, gene sets/transcription factors with names/symbols colored orange were significantly and consistently enriched in stroma in the LCM discovery and validation cohorts, whereas gene sets/transcription factors with names/symbols colored blue were consistently and significantly enriched in epithelium in the LCM discovery and validation cohorts (gene sets: adjusted p-value < 0.02; transcription factors: p < 0.05).