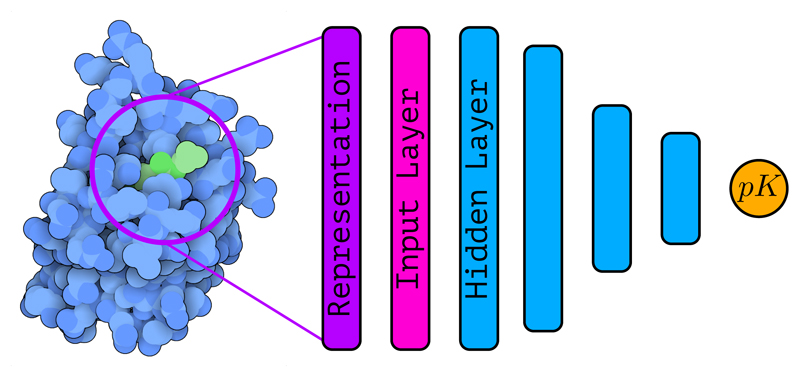
Figure 1.
Schematic representation of a structure-based deep learning architecture—with several hidden layers—for binding affinity prediction. The protein-ligand complex (PDB ID: 5S9H, rendered with Illustrate (Goodsell et al., 2019a)) is encoded into a suitable representation that is used as input to the deep learning architecture. Using a series of stacked hidden layers, a prediction of the binding affinity is finally obtained. The exact nature of the input layer as well as the hidden layers depends on the type of architecture under consideration.