Abstract
Individuals who have Down syndrome (DS) frequently develop early onset Alzheimer’s disease (AD), a neurodegenerative condition caused by the build-up of aggregated amyloid-β and tau proteins in the brain. Amyloid-β is produced by amyloid precursor protein (APP), a gene located on chromosome 21. People who have Down syndrome have three copies of chromosome 21 and thus also an additional copy of APP; this genetic change drives the early development of Alzheimer’s disease in these individuals. Here we use a combination of next-generation mouse models of Down syndrome (Tc1, Dp3Tyb, Dp(10)2Yey and Dp(17)3Yey) and a knockin mouse model of amyloid-β accumulation (AppNL-F) to determine how chromosome 21 genes, other than APP, modulate APP/amyloid-β in the brain when in three copies. Using both male and female mice, we demonstrate that three copies of other chromosome 21 genes are sufficient to partially ameliorate amyloid-β accumulation in the brain. We go on to identify a subregion of chromosome 21 that contains the gene/genes causing this decrease in amyloid-β accumulation and investigate the role of two lead candidate genes Dyrk1a and Bace2. Thus an additional copy of chromosome 21 genes, other than APP, can modulate APP/amyloid-β in the brain under physiological conditions. This work provides critical mechanistic insight into the development of disease and an explanation for the typically later age of onset of dementia in people who have ADDS, compared to those who have familial AD caused by triplication of APP.
Keywords: Amyloid precursor protein (APP), amyloid-β, Down syndrome, DYRK1A, BACE2
Introduction
DS is caused by trisomy of human chromosome 21 (Hsa21), and occurs in around 1/1000 live births in Europe (de Graaf, Buckley et al. 2021). Most individuals who have DS develop the neuropathological features of AD; amyloid-β plaques and tau neurofibrillary tangles by the age of 50 (Davidson, Robinson et al. 2018), and 80% of individuals will have developed dementia by age 65 (McCarron, McCallion et al. 2017). The high prevalence of AD in DS is in part due to the gene encoding amyloid precursor protein (APP) being located on Hsa21, thereby raising APP and amyloid-β protein levels (Glenner and Wong 1984, Cheon, Dierssen et al. 2008, Doran, Keator et al. 2017). Recent studies in preclinical systems have demonstrated that an extra copy of other genes on Hsa21 can modulate APP biology (Garcia-Cerro, Rueda et al. 2017, Wiseman, Pulford et al. 2018, Alic, Goh et al. 2020, Tosh, Rhymes et al. 2021) and thus may alter the earliest stages of AD in individuals who have DS. These extra genes may act to promote or to reduce amyloid-β accumulation, the mechanism that predominates is currently unclear. Notably, the age of clinical dementia diagnosis occurs slightly later in individuals who have DS, compared with those who have early-onset familial AD caused by duplication of APP (i.e., with three copies of wild-type APP) (Wiseman, Al-Janabi et al. 2015). However, a direct comparison between these two causes of AD is confounded by the different diagnostic criteria used (as necessitated by the underlying intellectual disability that occurs in people who have DS) (Benejam, Videla et al. 2020). Understanding these processes is crucial to the appropriate selection of treatments for AD-primary prevention trials in people who have DS.
Previous in vivo studies have either examined the processing of endogenous mouse APP or used APP transgenic models to address this biology, but both of these approaches have limitations (Garcia-Cerro, Rueda et al. 2017, Sasaguri, Nilsson et al. 2017, Wiseman, Pulford et al. 2018, Tosh, Rhymes et al. 2021). Mouse APP differs in sequence from the human protein. In the amyloid-β region these differences reduce both cleavage of the protein by β-secretase and the tendency of the amyloid-β generated to aggregate (Serneels, T’Syen et al. 2020), thus limiting our ability to determine how changes to biology affect accumulation of amyloid-β – a key early aspect of AD. The over- and mis-expression of APP in transgenic mouse models may cause artefactual phenotypes, masking the modulatory effect of the extra copy of Hsa21 genes and causing elevated mortality which may confound data interpretation (Saito, Matsuba et al. 2014, Sasaguri, Nilsson et al. 2017).
Here we take a combinatorial approach: assessing the effect of an additional copy of Hsa21 genes (using a series of DS mouse models (O’Doherty, Ruf et al. 2005, Yu, Li et al. 2010, Lana-Elola, Watson-Scales et al. 2016)) on the biology of endogenous mouse APP and on APP generated from a partially humanised App knock-in allele that also carries AD-causal Swedish (NL) and Iberian (F) point mutations, which does not cause elevated mortality (AppNL-F) (Fig. 1) (Saito, Matsuba et al. 2014). These data indicate that trisomy of genes on Hsa21 reduces amyloid-β accumulation and that people who have DS are partly protected from their raised APP gene dose by the additional copy of other genes on the chromosome. We go on to show that one of the gene or genes that cause this change in biology is located on mouse chromosome 16 between Mir802 and Zbtb21. This region contains 38 genes and we specifically test if mechanisms linked to two lead candidate genes in this region, Dyrk1a and Bace2, occur in our novel in vivo AD-DS model system.
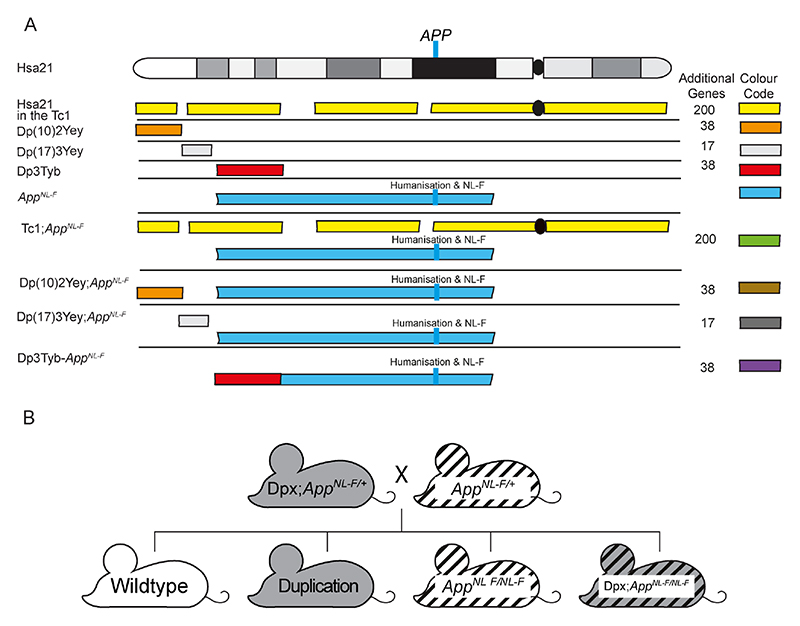
Figure 1. A schematic of Hsa21 indicating the major karyotypic bands, and the regions of Hsa21 or homologous regions of mouse chromosomes that are in three-copies in the mouse models used.
(A) The additional Hsa21 gene content in Tc1 (yellow), in the Dp(10)2Yey (orange), Dp(17)3Yey (light grey) and Dp3Tyb (red) segmental duplication models of DS as shown. The number of additional human genes in the Tc1, and the additional number of mouse genes in the Dp(10)2Yey, Dp(17)3Yey and Dp3Tyb as listed. These lines were crossed with the AppNL-F mouse model of amyloid-β accumulation (blue), to generate Tc1;AppNL-F(green), Dp(10)2Yey;AppNL-F (brown), Dp(17)3Yey;AppNL-F(dark grey), and Dp3Tyb-AppNL-F (purple) offspring, homozygous for the AppNL-F allele and carrying the additional gene content. The location of APP/App is indicated by the blue line, this locus is altered in the AppNL-F model such that the amyloid-β sequence is humanised and the model carries the AD-causal Swedish (NL) and Iberian (F) mutations. This colour scheme is used in subsequent figures to code for each of the mouse models. (B) Schematic of the generation of experimental cohorts for the duplication models (Dp(10)2Yey, Dp(17)3Yey and Dp3Tyb) used in the studies, experimental cohorts were produced by crossing Dpx;AppNL-F/+ mice with AppNL-F/+ mice.
Methods
Animal welfare and husbandry
All experiments were undertaken in accordance with the Animals (Scientific Procedures) Act 1986 (United Kingdom), after local institutional ethical review by the Medical Research Council, University College London and in accordance with ARRIVE2 guidelines (Percie du Sert, Hurst et al. 2020). Mice were housed in individually ventilated cages (Techniplast) with grade 5, autoclaved dust-free wood bedding, paper bedding and a translucent red “mouse house” at a specific pathogen free facility. Free-access to food (Picolab Rodent Diet 20 Labdiet) and water was provided. The animal facility was maintained at a constant temperature of 19-23°C with 55 ± 10% humidity in a 12 h light/dark cycle. Pups were weaned at 21 days and moved to standardised same-sex group housing with a maximum of 5 mice per cage.
The following mouse strains were used in this paper, here we show abbreviated name and then the official name and unique Mouse Genome Informatics (MGI) identifier: AppNL-F (Apptm2.1Tcs, MGI:5637816), Tc1 (Tc(HSA21)1TybEmcf, MGI:3814712), Dp3Tyb (Dp(16Mir802-Zbtb21)3TybEmcf, MGI:5703802), Dp(10)2Yey (Dp(10Prmt2-Pdxk)2Yey, MGI:4461400) and Dp(17)3Yey (Dp(17Abcg1-Rrp1b)3Yey, MGI:4461398).
Tc1 mice were maintained by mating Tc1 females to F1 (129S8 × C57BL/6) males. All other mouse strains were maintained by backcrossing males and females to C57BL/6J mice (imported from the Jackson Laboratory). Experimental cohorts for Tc1, Dp(10)2Yey and Dp(17)3Yey studies were produced by crossing mice carrying the additional Hsa21 or Hsa21 orthologous duplications with AppNL-F/+ animals in a two-generation cross to generate all required genotypes from the second generation (wild-type, Tc1/Dpx, AppNL-F/NL-F, Tc1/Dpx;AppNL-F/NL-F, Fig. 1, Table 1). As both the Dp3Tyb segmental duplication and the AppNL-F gene are located on mouse chromosome 16 (Mmu16), for this cross we first generated a Dp3Tyb-AppNL-F recombinant Mmu16, by crossing the two lines together and then back-crossing to C57BL/6J mice to identify recombined Mmu16, carrying both genetic changes on the same chromosome. Mice with the recombined Mmu16 were then crossed with AppNL-F/+ animals to generate Dp3Tyb;AppNL-F/NL-F progeny. For this cross AppNL-F/NL-F controls were generated from AppNL-F/+ x AppNL-F/+ matings, in addition to rare re-recombinations resulting in offspring without the Dp3Tyb segmental duplication but two copies of the App knock-in allele. Dp3Tyb controls were generated from Dp3Tyb x C57BL/6J matings generated at the same time as the Dp3Tyb;AppNL-F/NL-F mice. Wild-type (WT) controls were taken from all three matings.
Table 1. Summary of Mouse Cohorts.
Summary of the cohorts of mice, the cross(es) used to generate them, the age at which the cohort was sacrificed, the outcome measured and the figure in which the data are presented.
| Cohort | Crosses | Age | Outcomes | Figure | Female ‘n’ | Male ‘n’ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | Tc1/Dpx | App NL-F/NL-F | Tc1/Dpx; AppNL-F/NL-F | WT | Tc1/Dpx | App NL-F/NL-F | Tc1/Dpx; AppNL-F/NL-F | |||||
| 1 | Tc1;AppNL-F/+ and AppNL-F/+ | 3-months | APP and APP-CTF abundance | Figure 3 | 4 | 3 | 2 | 4 | 4 | 5 | 2 | 4 |
| 2 | Dp(10)2Yey;AppNL-F/+ and AppNL-F/+ | 3-months | APP and APP-CTF abundance | Figure 8 | 1 | 0 | 0 | 2 | 4 | 8 | 9 | 4 |
| 3 | Dp3Tyb-AppNL-F/+ and AppNL-F/+ Dp3Tyb and C57BL/6J AppNL-F/+ and AppNL-F/+ | 3-months | APP, APP-CTF, DYRK1A and BACE2 abundance Human amyloid-β fragments (only AppNLF carriers analysed) | Figure 7 Figure 10 & 11 | 6 | 5 | 8 | 5 | 11 | 7 | 7 | 8 |
| 4 | Dp3Tyb and C57BL/6J | 3-months | Mouse amyloid-β abundance | Figure 9 | 5 | 7 | N/A | N/A | 11 | 12 | N/A | N/A |
| 5 | Tc1;AppNL-F/+ and AppNL-F/+ | 8-months | 82E1 plaque counts Human amyloid-β abundance | Figure 2 | 5 | 5 | 15 | 8 | 0 | 3 | 8 | 10 |
| 6 | Dp3Tyb-AppNLF/+ and AppNL-F/+ AppNL-F/+ and AppNL + | 8-months | 82E1 plaque counts Human amyloid-β abundance | Figure 4 Figure 5 & 6 | 2 | Not available because of breed scheme | 2 | 9 | 2 | Not available because of breed scheme | 7 | 5 |
| 7 | Dp(10)2Yey;AppNL-F/+ and AppNL-F/+ | 8-months | 82E1 plaque counts Human amyloid-β abundance | Figure 4 Figure 5 & 6 | 2 | 2 | 4 | 6 | 4 | 1 | 5 | 3 |
| 8 | Dp(17)3Yey; AppNL-F/+ and AppNL-F/+ | 8-months | 82E1 plaque counts Human amyloid-β abundance | Figure 4 Figure 5 & 6 | 3 | 3 | 7 | 7 | 3 | 3 | 5 | 7 |
Animals were euthanized by exposure to rising carbon dioxide, followed by confirmation of death by dislocation of the neck in accordance with the Animals (Scientific Procedures) Act 1986 (United Kingdom).
Tissue preparation and western blotting
For analysis of protein abundance in the hippocampus and cortex, tissues were dissected under ice cold phosphate-buffered saline (PBS) before snap freezing. Samples were then homogenised in RIPA Buffer (150 mM sodium chloride, 50 mM Tris, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) plus complete protease inhibitors (Calbiochem) by mechanical disruption. Total protein content was determined by Bradford assay or Pierce 660 nm assay (Thermo Fisher). Samples from individual animals were analysed separately and were not pooled.
Equal amounts of total brain proteins were then denatured in LDS denaturing buffer (Invitrogen) and β-mercaptoethanol, prior to separation by SDS-PAGE gel electrophoresis using precast 4-12% Bis-Tris gels (Invitrogen). Proteins were transferred to nitrocellulose or PVDF membranes prior to blocking in 5% milk/PBST (0.05% Tween 20) or 5-10% bovine serum albumin (BSA)/PBST. Primary antibodies were diluted in 1% BSA/PBST, HRP-conjugated secondary anti-rabbit, anti-mouse and anti-goat antibodies (Dako) were diluted 1:10,000 in 1% BSA/PBST. Linearity of antibody binding was confirmed by a 2-fold dilution series of cortical protein samples. Band density was analysed using Image J. Relative signal of the antibody of interest compared to the internal loading control was then calculated, and the relative signal was then normalized to the mean relative signal of control samples electrophoresed on the same gel. Means of technical replicates were calculated and used for ANOVA, such that biological replicates were used as the experimental unit.
Primary antibodies against C-terminal APP (Sigma A8717, 1:10,000), β-actin (Sigma A5441, 1:60,000), DYRK1A (7D10, Abnova, 1:500) and BACE2 (Abcam ab5670 1:1,000) were used.
Biochemical fractionation of mouse brain tissues for the analysis of human amyloid-β
Cortical proteins were fractionated as described in Shankar et al. (2009). A half cortex was weighed on a microscale and homogenised in 4 volumes of ice-cold Tris-buffered saline (TBS) (50mM Tris-HCl pH 8.0) containing a cocktail of protease and phosphatase inhibitors (Calbiochem) using a handheld mechanical homogeniser and disposable pestles (Anachem). Samples were then transferred to 1.5 ml microfuge tubes (Beckman Coulter #357448), balanced by adding more TBS and centrifuged at 175,000 × g with an RC-M120EX ultracentrifuge (Sorvall) fitted with rotor S100AT5 at 4 °C for 30 mins. Supernatant (the Tris- fraction) was removed and stored at -80 °C. The remaining pellet was homogenised in 5 volumes of ice-cold 1% Triton X-100 (Sigma-Aldrich) in TBS (50mM Tris-HCl pH 8.0), balanced and centrifuged at 175,000 × g for 30 mins at 4 °C. The resultant supernatant (the Triton-soluble fraction) was removed and stored at -80 °C. The pellet was then re-suspended in 8 volumes (by original cortical weight) of TBS (50mM Tris-HCl pH 8.0), containing 5 M guanidine HCl and left overnight at 4 °C on a rocker to ensure full re-suspension, and subsequently stored at -80 °C. A Bradford assay or Pierce 660 nm protein assay (Thermo Fisher) was performed to determine protein concentration.
Biochemical preparation of mouse brain tissues for the analysis of mouse amyloid-β
A half cortex was weighed on a microscale and homogenised in 3 volumes of ice-cold TBS (50mM Tris-HCl pH 8.0) containing a cocktail of protease and phosphatase inhibitors (Calbiochem) using a handheld mechanical homogeniser and disposable pestles (Anachem) based on the method in reference (Holtta, Hansson et al. 2013). Homogenates were centrifuged at 21130 × g at 4 °C for 1 hour and the resultant supernatant was stored at -80 °C for onward analysis.
Quantification of Aβ abundance by Meso Scale Discovery Assay
Amyloid-β38, amyloid-β40 and amyloid-β42 levels were quantified on Multi-Spot 96 well plates pre-coated with anti-amyloid-β38, amyloid-β40 and amyloid-β42 antibodies using multiplex MSD technology, as described in (Wiseman, Pulford et al. 2018). A 6E10 detection antibody was used to quantify human amyloid-β and 4G8 detection antibody for the quantification of mouse amyloid-β. Amounts of amyloid-β38, amyloid-β40 and amyloid-β42 were normalised to the original starting weight of cortical material.
Immunohistochemistry
Half brains were immersion fixed in 10% buffered formal saline (Pioneer Research Chemicals) for a minimum of 48 hours prior to being processed to wax (Leica ASP300S tissue processor). The blocks were trimmed laterally from the midline by ~0.9-1.4 mm to give a sagittal section of the hippocampal formation. Two 4 μm sections at least 40 μm apart were analysed. The sections were pre-treated with 98% formic acid for 8 mins, followed by washing. The slides were wet loaded onto a Ventana XT for staining (Ventana Medical Systems, Tucson, AZ, USA). The protocol included the following steps: heat induced epitope retrieval (mCC1) for 30 mins in Tris Boric acid EDTA buffer (pH 9.0), superblock (8 mins) and manual application of 100 μl directly biotinylated mouse monoclonal IgG1 antibody against the N-terminus of Aβ (82E1, IBL, 0.2 μg/ml) for 8 hours. Staining was visualised using a ChromoMap DAB kit followed by counterstaining with haematoxylin. The sections were dehydrated, cleared, and mounted in DPX prior to scanning (Leica SCN400F slide scanner). All images were analysed using ImageJ and by manual plaque counting by two independent researchers.
Biochemical preparation of mouse brain tissues for mass spectrometry of amyloid-β
A half cortex was weighed on a microscale and was homogenized in 5 volumes of tris(hydroxymethyl)aminomethane (Tris)-buffered saline (TBS), pH 7.6, containing cOmplete™ Protease Inhibitor Cocktail (Roche, cat: 04693116001). For the homogenization, one 5 mm bead per sample was used in a TissueLyser (Qiagen) for 4 min at 30 Hz. After homogenization, additional TBS with protease inhibitor cocktail was added up to 1 ml and transferred to a new tube to be centrifuged at 31,000 g for 1 h at 4 °C. The pellet was resuspended in 1 ml of 70% formic acid (FA) (v/v), followed by further homogenization in the TissueLyser for 2 min at 30 Hz and subsequent sonication for 30 s. The homogenate was centrifuged again at 31,000 g for 1 h at 4 °C and the supernatant (FA fraction) was dried down in a vacuum centrifuge.
Initially, 400 μl of 70% FA (v/v) was added to the dried FA fractions, shaken for 30 min at 21°C and centrifuged at 31,000 g for 1 h at4 °C. After the removal of the supernatant, neutralization with 8 ml 0.5 M Tris was performed. Immunoprecipitation (IP) was performed as previously described with some modifications (Gkanatsiou, Portelius et al. 2019). Briefly, 50 μl of sheep anti-mouse magnetic beads (Thermo Fisher Scientific) that had previously been linked with 4 μg each of mouse monoclonal 6E10 and 4G8 antibodies (Biolegend) were added to the neutralized FA fraction. This complex was incubated overnight at 4 °C in 0.2% Triton X-100 in PBS (v/v). By using an automated magnetic-particle KingFisher ml system (Thermo Fisher Scientific), the samples were then washed with PBS Triton X-100, PBS, and 50 mM ammonium bicarbonate separately before elution in 100 μl 0.5% FA. Eluates were dried down in a vacuum centrifuge and stored at -80 °C pending mass spectrometry (MS) analysis.
Mass spectrometry
Liquid chromatography-mass spectrometry (LC-MS) was conducted in a similar manner as described previously (Portelius, Tran et al. 2007). Briefly, a nanoflow liquid chromatograph was coupled to an electrospray ionization (ESI) hybrid quadrupole–orbitrap tandem MS (Dionex Ultimate 3000 system and Q Exactive, both Thermo Fisher Scientific). Samples were reconstituted in 7 μl 8% FA/8% acetonitrile in water (v/v/v) and loaded onto an Acclaim PepMap 100 C18 trap column (length 20 mm; inner diameter 75 μm; particle size 3 μm; pore size 100 Å) for online desalting, and thereafter separated on a reversed-phase Acclaim PepMap RSLC column (length 150 mm, inner diameter 75 μm; particle size 2 μm; pore size 100 Å) (both Thermo Fisher Scientific). Mobile phases were A: 0.1% FA in water (v/v), and B: 0.1% FA/84% acetonitrile in water (v/v/v). The flow rate was 300 nl/min and a linear gradient of 3-40% B for 50 min was applied. The temperature of the column oven was 60 °C. Mass spectrometer settings were as follows: positive ion mode; mass-to-charge (m/z) interval 350-1800 m/z units; data dependent acquisition with 1 precursor ion acquisition (MS) followed by up to 5 fragment ion acquisitions (MS/MS); resolution setting 70,000 (for both MS and MS/MS); number of microscans 1 (MS and MS/MS); target values 106 (MS and MS/MS); maximum injection time 250 ms (MS and MS/MS); fragmentation type was higher-energy collisional dissociation fragmentation (HCD); normalised collision energy (NCE) setting 25; singly charged ions and ions with unassigned charge were excluded for MS/MS selection. Database search (including isotope and charge deconvolution) and label free quantification was performed with PEAKS Studio v8.5 (Bioinformatics Solutions Inc.) against a custom-made APP database. All suggested fragment mass spectra were evaluated manually.MS signal was normalised to starting weight of the cortex prior to data analysis.
Statistical analysis and experimental design
All experiments and analyses were undertaken blind to genotype and sex. A 6-digit identification number was allocated to each animal prior to genotyping which was used to blind samples. Experimental groups for all experiments were pseudorandomised using Mendelian inheritance. Mice not carrying the correct combination of alleles (i.e. AppNL-F/+ heterozygous mice) were excluded from the analysis, no other animals were excluded. Some samples were lost from the study because of technical issues during tissue processing to wax or during fractionation. Data were analysed as indicated in figure legends by univariate ANOVA with between-subject factors being genetic status (App+/+/AppNL-F/NL-F and/or wild-type/Hsa21 or wild-type/Dpx) and sex. Fractionation batch was included as a between-subject factor for analysis of amyloid-β by MSD assays. The subject means of technical replicates were calculated and used in the ANOVA for western blot and MSD assays, as the number of replicates for which data was available varied between samples. Repeat measures ANOVA was used for manual plaque counts (combining the data of two independent researchers).
Results
Trisomy of chromosome 21 decreases accumulation of amyloid-β in the cortex of the AppNL-F mouse model
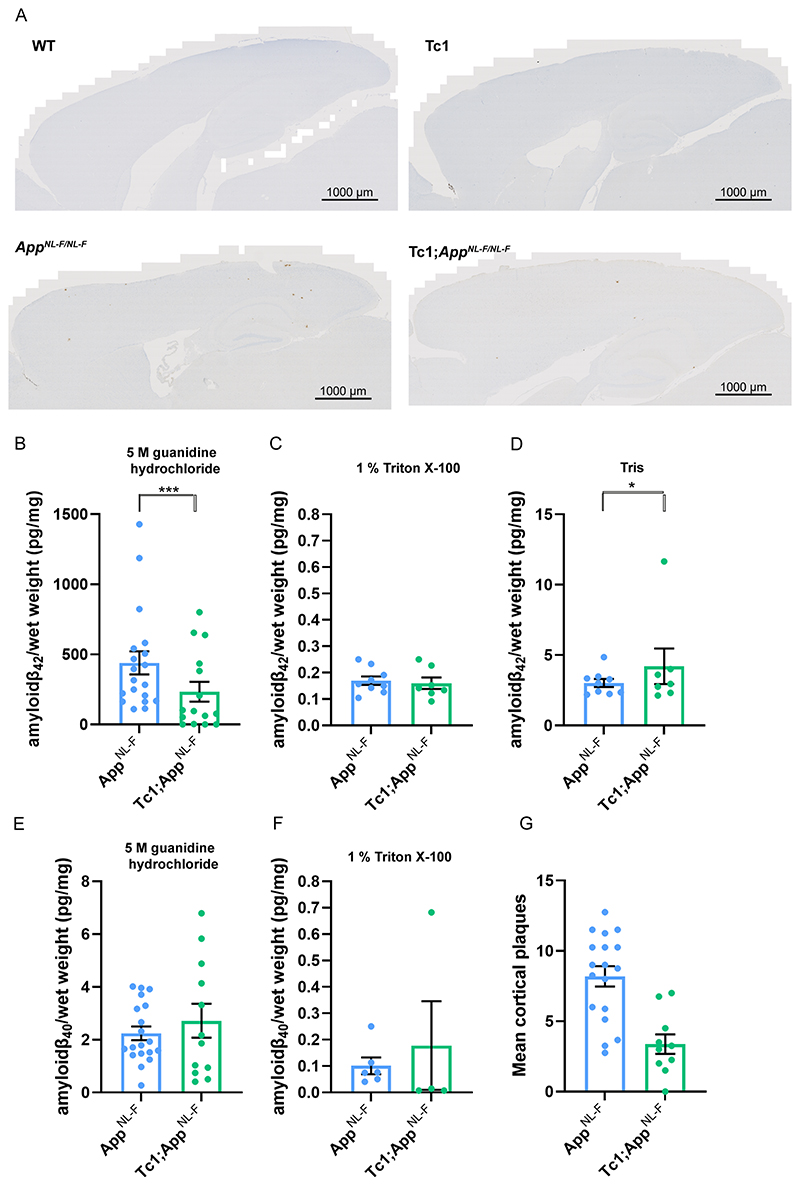
Following on from our previous studies, which indicated that an additional copy of Hsa21 alters APP biology and the accumulation of amyloid-β in vivo in an APP transgenic model (Wiseman, Pulford et al. 2018, Tosh, Rhymes et al. 2021), here we determined if an additional copy of Hsa21 modulated APP/amyloid-β biology in the AppNL-F knock-in mouse model. We undertook a two generation cross of the Tc1 mouse model of DS (O’Doherty, Ruf et al. 2005), which contains a freely segregating copy of Hsa21 (but not a functional additional copy of APP) (Gribble, Wiseman et al. 2013), with the AppNL-F model to generate 4 genotypes of mice (wild-type, Tc1, AppNL-F/NL-F and Tc1;AppNL-F/NL-F). We quantified the number of 82E1+ amyloid-β deposits in the cortex of these 4 genotypes of mice at 8-months of age. 82E1+ deposits were not observed in wild-type or Tc1 mice consistent with our previous study (Wiseman, Pulford et al. 2018). We found a significant decrease in the number of deposits in Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (Fig. 2 A, G).
Figure 2. Trisomy of Hsa21 results in a decrease in amyloid-β deposition in the AppNL-F/NL-F model.
(A, G) Amyloid-β deposition (82E1) in the cortex was quantified at 8-months of age in male and female mice by manual plaque counting. (A) Representative image of 82E1 stained amyloid-β deposits (brown) in wild-type (WT), Tc1, AppNL-F/NL-F, Tc1;AppNL-F/NL-F mice. (G) Significantly fewer amyloid-β deposits in the cortex were observed in Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,23) = 24.997, p < 0.001). AppNL-F/NL-F female n = 11, male n = 7, Tc1;AppNL-F/NL-F female n = 4, male n = 6. No amyloid-β deposits were observed in WT (female n = 5) or Tc1 (female n = 5, male n = 3) age matched littermate controls (data not shown). (B-F) Total cortical proteins were biochemically fractionated and amyloid abundance analysed by MSD assay. (B) No statistically significant difference in the abundance of 5 M guanidine hydrochloride soluble amyloid-β42 was observed between Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,13) = 2.262, p = 0.156). AppNL-F/NL-F female n = 15, male n = 8; Tc1;AppNL-F/NL-F female n = 8, male n = 10. WT and Tc1 samples which do not produce human amyloid-β were included as negative controls. WT n = 4, of which 1 was below limit of detection ( pg/mg, SEM = 0.433); Tc1 n = 6, of which 4 samples were below the limit of detection ( pg/mg, SEM 1.437). (C) No difference in the abundance of 1% Triton X-100 soluble amyloid-β42 was observed between Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,5) = 0.015, p = 0.907). AppNL-F/NL-F female n = 7, male n = 2 (n = 8 below limit of detection); Tc1;AppNL-F/NL-F female n = 5, male n = 2 (n = 11 below limit of detection). WT and Tc1 samples which do not produce human amyloid-β were included as negative controls. WT n = 4, of which 2 were below the limit of detection ( pg/mg, SEM = 0.007); Tc1 n = 6, of which all samples were below the limit of detection. (D) Significantly more Tris soluble amyloid-β42 was observed in Tc1; AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,5) = 10.697, p = 0.022). AppNL-F/NL-F female n = 7, male n = 2 (n = 8 below limit of detection) Tc1;AppNL-F/NL-F female n = 5, male n = 2 (n = 11 below limit of detection). WT and Tc1 samples which do not produce human amyloid-β were included as negative controls. WT n = 4, of which all were below the limit of detection; Tc1 n = 6, of which all samples were below the limit of detection. (E) No difference in the abundance of 5 M guanidine hydrochloride-soluble amyloid-β40 was observed between Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,13) = 0.005, p = 0.946). AppNL-F/NL-F female n = 11, male n = 8, n = 4 below the limit of detection; Tc1;AppNL-F/NL-F female n = 7, male n = 5, n = 6 below the limit of detection. WT and Tc1 samples which do not produce human amyloid-β were included as negative controls. WT n = 4, of which 3 were below the limit of detection (1.870 pg/mg); Tc1 n = 6, of which 5 samples were below the limit of detection (1.150 pg/mg). (F) No difference in the abundance of 1% Triton X-100-soluble amyloid-β40 was observed between Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F control (F(1,3) = 0.000, p = 0.991). AppNL-F/NL-F female n = 4, male n = 0 (n = 17 below the limit of detection); Tc1;AppNL-F/NL-F female n = 4, male n = 2 (n = 14 below the limit of detection). Tris-soluble amyloid-β40 was not detected in these samples. WT and Tc1 samples which do not produce human amyloid-β were included as negative controls. WT n = 4, of which 2 samples were below limit of detection ( pg/mg, SEM = 0.005); Tc1 n = 6, of which all samples were below the limit of detection. Error bars show SEM, data points are independent mice.
We also determined if trisomy of Hsa21 modulated the biochemical aggregation of amyloid-β40 and amyloid-β42 in the cortex at 8-months of age, using biochemical protein fractionation by step-wise homogenisation and ultracentrifugation in sequentially more disruptive solutions (Tris-HCl, Tris-HCl 1% Triton X-100 and finally 5 M guanidine hydrochloride). We then quantified human amyloid-β40 and amyloid-β42 in each fraction normalised to starting brain weight (6E10 MSD triplex assay). Samples from mice without a humanised App allele, that do not produce human amyloid-β, were used as negative controls. Amyloid-β42 in the guanidine hydrochloride fraction was not significantly reduced in the presence of the extra copy of Hsa21 (Fig. 2B). A significant increase in Tris-soluble amyloid-β42 was seen in the cortex of Tc1;AppNL-F/NL-F compared with AppNL-F/NL-F controls (Fig. 2D). However, we note that this effect is being driven by one outlier and significance is lost if this animal is excluded from the analysis moreover this analyte could not be detected in a significant proportion of the samples analysed. Thus a replication study is required to determine the validity of this result. No significant difference in Triton-soluble amyloid-β42 abundance was observed (Fig. 2C). The amount of human amyloid-β40 in the AppNL-F/NL-F model is very low because of the Iberian mutation in the modified App allele and this analyte was below the limit of detection in the Tris-soluble fraction and did not significantly differ between genotypes in the Triton and 5 M guanidine hydrochloride fractions (Fig. 2E, F). These data indicate that an additional copy of a Hsa21 gene or genes is sufficient to reduce the deposition of amyloid-β in the cortex, this may occur via an effect on amyloid-β formation, clearance or aggregation,
Decreased amyloid-β accumulation in the Tc1-AppNL-F/NL-F model does not occur because of a reduction of APP or CTF-β abundance
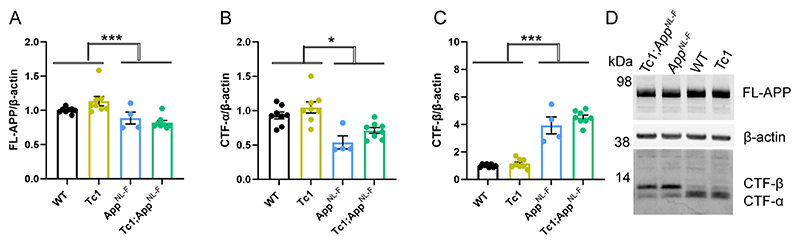
The significant increase in Tris-soluble amyloid-β42 in the Tc1;AppNL-F/NL-F cortex suggests that the decrease in amyloid-β accumulation observed is likely to be caused by a change in peptide aggregation. However, previous studies have suggested that genes on Hsa21, other than APP, can increase APP protein level in vivo and modulate the abundance of the amyloid-β precursor APP-C-terminal fragment-β (CTF-β) (Garcia-Cerro, Rueda et al. 2017, Naert, Ferre et al. 2018, Wiseman, Pulford et al. 2018). Thus, we used western blotting to determine if the additional Hsa21 genes altered APP or CTF-β abundance in our new model system. Here, we found no evidence of decreased mouse or human full-length APP (FL-APP) in the cortex of the Tc1 and Tc1;AppNL-F/NL-F compared with wild-type and AppNL-F/NL-F mice (Fig. 3A, D). We note that significantly less FL-APP is detected in humanised models compared with controls (using antibody A8717), this may reflect a reduction in antibody binding rather than a biological reduction in protein level.
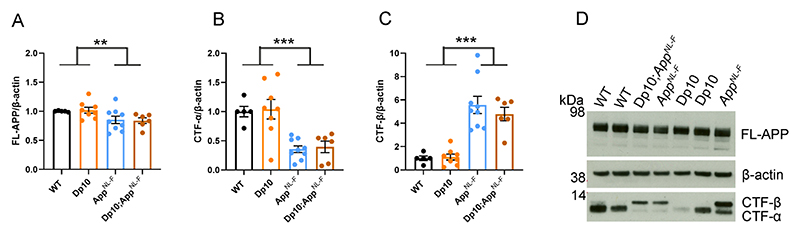
Figure 3. The abundance of FL-APP and CTF is not altered by trisomy of Hsa21.
(A-D) The relative abundance of full-length APP (FL-APP), APP β-C-terminal fragment (β-CTF) and APP α-C-terminal fragment (α-CTF) compared to β-actin was measured by western blot using A8717 primary antibody in the cortex at 3-months of age in female and male mice. (A) Significantly less FL-APP was observed in mice in which App was humanised and mutated (F(1,19) = 23.837, p < 0.001). An additional copy of Hsa21 did not alter FL-APP abundance (F(1,19) = 0.599, p = 0.449). (B) Significantly less CTF-α was observed in mice in which App was humanised and mutated (F(1,19) = 5.950, p = 0.025) but an additional copy of Hsa21 did not alter α-CTF abundance (F(1,19) = 3.012, p = 0.099). (C) Significantly more β-CTF was observed in mice in which App was humanised and mutated (F(1,19) = 868.431, p < 0.001). By ANOVA a significant effect of Hsa21 on CTF-β abundance was detected (F(1,19) = 23.462, p < 0.001), however wild-type (WT) and Tc1 (Bonferroni pair-wise comparison p = 1.000) and AppNL-F/NL-F and Tc1;AppNL-F/NL-F (Bonferroni pair-wise comparison p = 0.118) were not statistically significant. WT female n = 4, male n = 4: Tc1 female n = 3, male n = 5; AppNL-F/NL-F female n = 2, male n = 2; Tc1;AppNL-F/NL-F female n = 4, male n = 4. (D) Representative image of western blot in WT, Tc1, AppNL-F/NL-F, Tc1;AppNL-F/NL-F mice. Error bars show SEM, data points are independent mice.
We found a significant increase in CTF-β and a significant decrease in CTF-α in the cortex of AppNL-F/NL-F mice consistent with the reported effects of the introduced mutations on APP processing (Fig. 3B, C, D) (Saito, Matsuba et al. 2014). An additional copy of Hsa21 did not significantly alter wild-type APP CTF-β or CTF-α levels in the cortex of the Tc1 compared to wild-type-mice or in the Tc1;AppNL-F/NL-F compared to AppNL-F/NL-F. This finding contrasted with the large increase in CTF-β in male Tc1;APP transgenic model that we previously reported (Wiseman, Pulford et al. 2018). These data suggest that the reduction in deposition of amyloid-β in the Tc1;AppNL-F/NL-F model is likely mediated by an enhancement of amyloid-β clearance or an impairment of peptide aggregation rather than a decrease in APP or CTF-β abundance.
Decreased accumulation of amyloid-β is caused by an additional copy of 38 Hsa21 orthologous genes
Many DS-associated phenotypes are multigenic, caused by the combined effect of multiple Hsa21 genes acting together on one biological pathway. To understand the mechanisms underlying the decrease in amyloid-β accumulation in the Tc1;AppNL-F/NL-F model further, we used a series of mouse models of DS that carry an extra copy of subregions of mouse chromosomes that are orthologous with Hsa21, to identify the combination of regions/genes responsible for the changes (Fig. 1) (Yu, Li et al. 2010, Lana-Elola, Watson-Scales et al. 2016). We used 3 mouse lines that carry similar gene content to the Tc1 mouse model. However, because of the limitations of available models we were unable to explore the Hsa21 genes closest to App on Mmu16, which are in three copies in the Tc1 model, as using recombination to generate the required combination of alleles in the available Dp9Tyb model was not feasible.
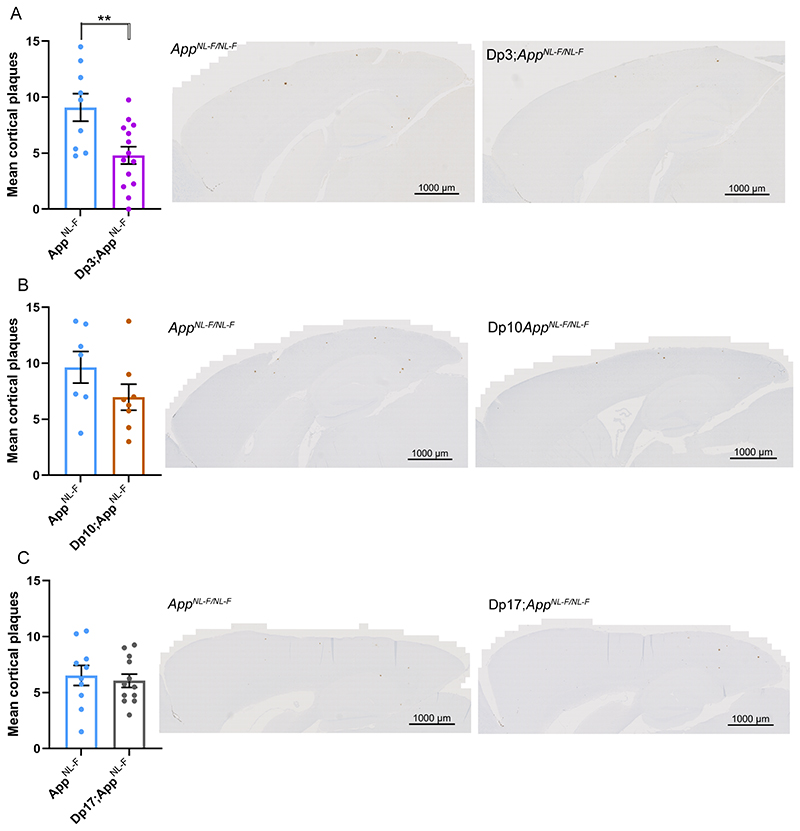
An extra copy of the genes between Mir802 and Zbtb21 (Dp3Tyb) was sufficient to decrease the accumulation of amyloid-β in the cortex, as quantified by 82E1 plaque counts (Fig. 4A). However, an extra copy of the genes between Abcg1 and Rrp1b (Dp(10)2Yey) and between Prmt2 and Pdxk (Dp(17)3Yey) was not sufficient to significantly alter the accumulation of amyloid-β in the cortex, as quantified by 82E1 plaque counts in 8-month old mice (Fig. 4B, C).
Figure 4. Duplication of the Dp3Tyb region of mouse chromosome 21 orthologous with Hsa21 is sufficient to decrease amyloid-β accumulation in the AppNL-F/NL-F model.
(A-C) Amyloid-β deposition (82E1) in the cortex was quantified at 8-months of age in male and female mice by manual plaque counting. (A) Significantly fewer amyloid-β deposits were observed in the cortex of Dp3Tyb;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,18) = 12.359, p = 0.002). AppNL-F/NL-F female n = 2, male n = 7; Dp3Tyb;AppNL-F/NL-F female n = 9, male n = 5. No amyloid deposits were observed in n = 4 WT controls. (B) A non-significant trend for reduced amyloid-β deposits in the cortex was observed in Dp(10)2Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,11) = 12.359, p = 0.077). AppNL-F/NL-F female n = 4, male n = 3; Dp(10)2Yey;AppNL-F/NL-F female n = 6, male n = 2. No amyloid-β deposits were observed in WT n = 6 and Dp(10)2Yey n = 3 controls. (C) No difference in amyloid-β deposits were observed in the cortex of Dp(17)3Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,18) = 0.021, p = 0.885). AppNL-F/NL-F female n = 6, male n = 4; Dp(17)3Yey AppNL-F/NL-F female n = 6, male n = 6. No amyloid-β deposits were observed in WT n = 6 and Dp(17)3Yey n = 6 controls. Dp3Tyb abbreviated to Dp3, Dp(10)2Yey abbreviated to Dp10, Dp(17)3Yey abbreviated to Dp17 for clarity. Error bars show SEM, data points are independent mice
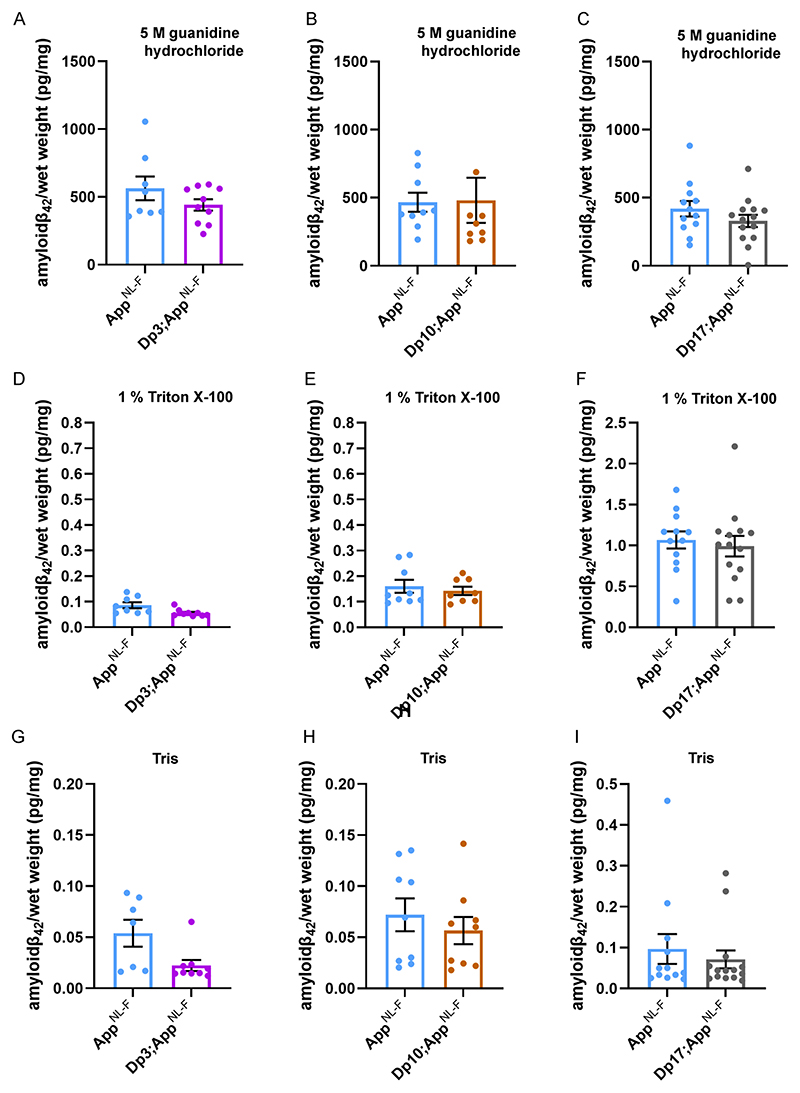
The abundance of amyloid-β42 in the guanidine hydrochloride fraction was not significantly altered in the Dp3Tyb;AppNL-F/NL-F, Dp(10)2Yey;AppNL-F/NL-F or
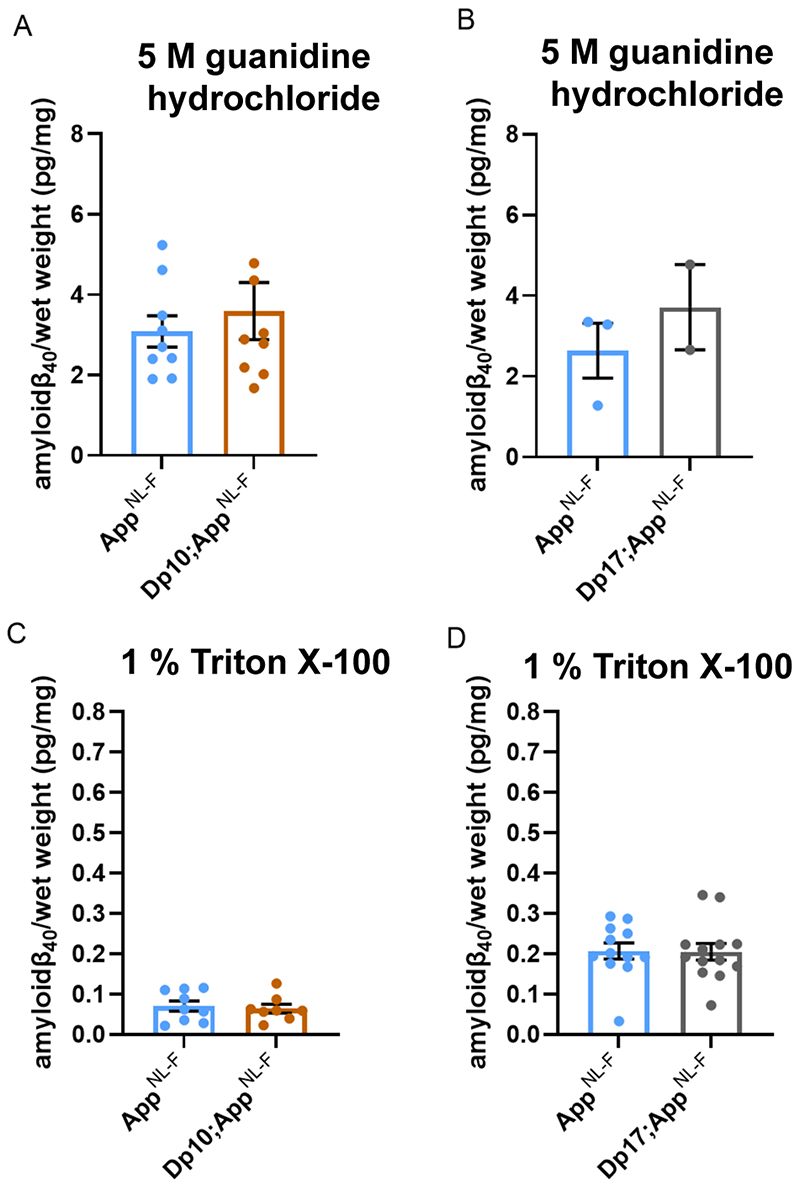
Dp(17)3Yey;AppNL-F/NL-F mice compared with AppNL-F/NL-F controls at 8-months of age (Fig. 5A-C). No significant difference in the abundance of Tris- or Triton-soluble amyloid-β42 was observed in Dp3Tyb;AppNL-F/NL-F, Dp(10)2Yey;AppNL-F/NL-F or Dp(17)3Yey;AppNL-F/NL-F compared to controls (Fig. 5D-I). No difference in the abundance of guanidine hydrochloride or Triton-soluble amyloid-β40 was observed between Dp(10)2Yey;AppNL-F/NL-F or Dp(17)3Yey;AppNL-F/NL-F compared to controls (Fig. 6A-D). These data indicate that a gene or genes in 3-copies in the Dp3Tyb model is sufficient to decrease the deposition of amyloid-β in the brain.
Figure 5. Biochemical solubility of amyloid-β42 is not altered by an additional copy of the Dp3Tyb, Dp(10)2Yey or Dp(17)3Yey Hsa21 orthologous regions.
Total cortical proteins were biochemically fractionated from 8-month of age mice and amyloid-β abundance analysed by MSD assay (6E10). No difference in the abundance of (A) 5 M guanidine hydrochloride soluble amyloid-β42 (F(1,12) = 0.128, p = 0.726), (D) 1% Triton X-100 soluble amyloid-β42 (F(1,12) = 2.863, p = 0.116) or (G) Tris-soluble amyloid-β42 (F(1,10) = 0.281, p = 0.608) was observed between Dp3Tyb; AppNL-F/NL-F compared with AppNL-F/NL-F controls. AppNL-F/NL-F female n = 2, male n = 6; Dp3Tyb;AppNL-F/NL-F female n = 7, male n = 3. No difference in the abundance of (B) 5 M guanidine hydrochloride soluble (F(1,11) = 2.337, p = 0.115), (E) 1% Triton X-100-soluble amyloid-β42 (F(1,11) = 0.145, p = 0.711) or (H) Tris-soluble amyloid-β42 (F(1,11) = 0.001, p = 0.978) was observed between Dp(10)2Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls. AppNL-F/NL-F female n = 4, male n = 5; Dp(10)2Yey;AppNL-F/NL-F female n = 6, male n = 3. No difference in the abundance of (C) 5 M guanidine hydrochloride soluble (F(1,20) = 4.079, p = 0.057), (F) 1% Triton X-100 soluble amyloid-β42 (F(1,20) = 0.124, p = 0.728) or (I) Tris soluble (F(1,20) = 0.521, p = 0.479) amyloid-β42 was observed between Dp(17)3Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls. AppNL-F/NL-F female n = 7, male n = 5; Dp(17)3Yey;AppNL-F/NL-F female n = 7, male n = 7. Details of negative controls, which do not carry an AppNL-F allele in Table 2. Dp3Tyb abbreviated to Dp3, Dp(10)2Yey abbreviated to Dp10, Dp(17)3Yey abbreviated to Dp17 for clarity. Error bars show SEM, data points are independent mice.
Figure 6. Biochemical solubility of amyloid-β40 is not altered by an additional copy of the Dp(10)2Yey or Dp(17)3Yey Hsa21 orthologous regions.
Total cortical proteins were biochemically fractionated from 8-month of age mice and amyloid-β abundance analysed by MSD assay (6E10). (A) No difference in the abundance of 5 M guanidine hydrochloride-soluble amyloid-β40 was observed between Dp(10)2Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,12) = 2.137, p = 0.169). AppNL-F/NL-F female n = 6, male n = 3; Dp(10)2Yey;AppNL-F/NL-F female n = 4, male n = 5. (B) No difference in the abundance of 5 M guanidine hydrochloride-soluble amyloid-β40 was observed between Dp(17)3Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,1) = 0.782, p = 0.539). AppNL-F/NL-F female n = 1, male n = 2, n = 9 below limit of detection; Dp(17)3Yey;AppNL-F/NL-F female n = 2, male n = 0, n = 12 below limit of detection. (C) No difference in the abundance of 1% Triton X-100-soluble amyloid-β40 was observed between Dp(10)2Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,11) = 0.540, p = 0.478). AppNL-F/NL-F female n = 6, male n = 3; Dp(10)2Yey;AppNL-F/NL-F female n = 4, male n = 4. (D) No difference in the abundance of 1% Triton X-100-soluble amyloid-β40 was observed between Dp(17)3Yey;AppNL-F/NL-F compared with AppNL-F/NL-F controls (F(1,19) = 0.007, p = 0.982). AppNL-F/NL-F female n = 7, male n = 5; Dp(17)3Yey;AppNL-F/NL-F female n = 7, male n = 6, n = 1 below limit of detection. Details of negative controls, which do not carry an AppNL-F allele in Table 2. Error bars show SEM, data points are independent mice. Dp(10)2Yey abbreviated to Dp10 and Dp(17)3Yey abbreviated to Dp17 for clarity.
Increased DYRK1A does not lead to increased APP or amyloid-β in the Dp3Tyb model of DS
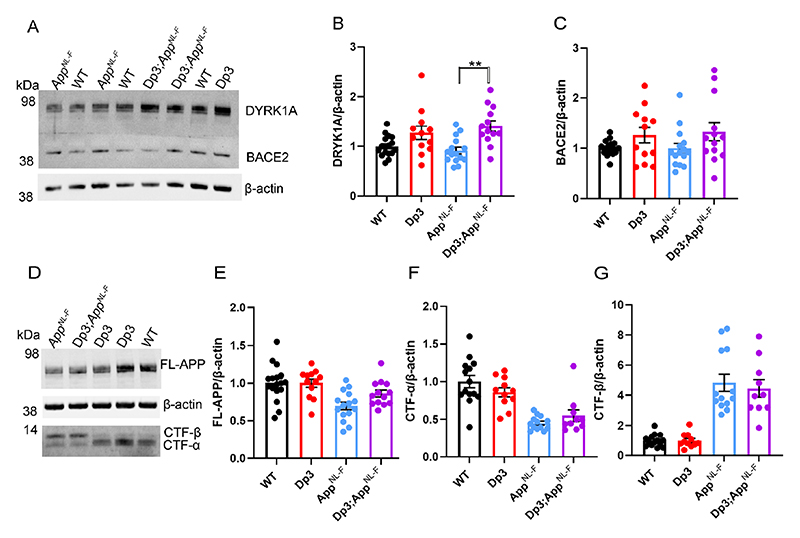
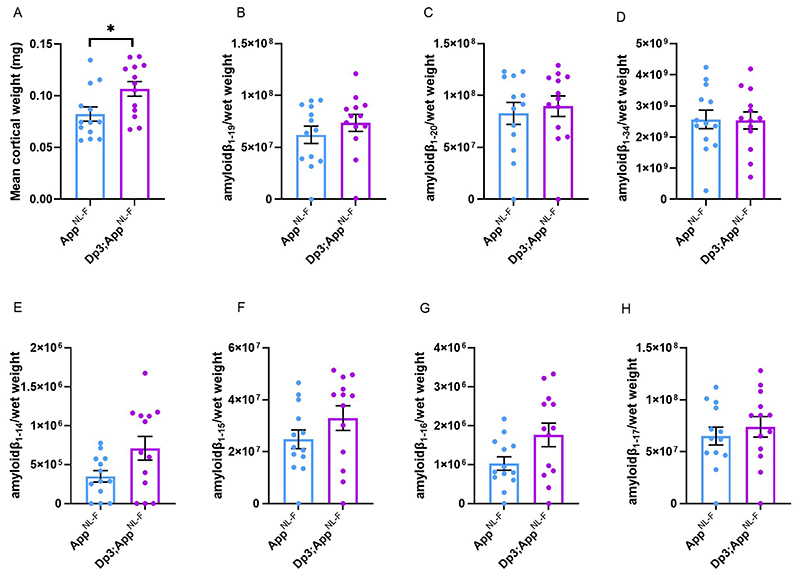
The Dp3Tyb model contains an additional copy of 38 genes, including Dyrk1a, a kinase that phosphorylates APP (Ryoo, Cho et al. 2008), increasing the abundance of the protein in vivo, contributing to raised soluble amyloid-β abundance in the Ts65Dn mouse model of DS (Garcia-Cerro, Rueda et al. 2017). We wanted to determine whether we could observe this previously reported biology in our new model system. We found that an extra copy of the Dp3Tyb region raises the abundance of DYRK1A in the cortex of 3-month-old animals, including in the context of AppNL-F knock-in mutations (Fig. 7A-B). This is consistent with numerous previous reports of dosage sensitivity of DYRK1A in the mouse throughout lifespan (Sheppard, Plattner et al. 2012, Garcia-Cerro, Rueda et al. 2017, Yin, Jin et al. 2017). We also note that consistent with previous reports in other mouse model systems, 3-copies of Dyrk1a in the Dp3Tyb;AppNL-F/NL-F model was associated with an increase in cortical weight (Guedj, Pereira et al. 2012) (Fig. 10A).
Figure 7. Duplication of the Dp3Tyb region is sufficient to raise the protein abundance of DYRK1A in the cortex but does not significantly alter BACE2, FL-APP or CTF abundance.
The abundance of (A-B) DYRK1A, (A, C) BACE2, (D, E) full-length, APP (FL-APP), (D, F) C-terminal fragment-α (CTF-α) and (D, G) C-terminal fragment-β (CTF-β) relative to β-actin loading control was measured by western blot in the cortex at 3 months of age in male and female mice. ANOVA analysis indicated that an additional copy of the Dp3Tyb region increased the abundance of (B) DYRK1A (F(1,49) = 16.511, p < 0.001) and (C) BACE2 F(1,49) = 4.444, p = 0.040). Post-hoc pair-wise comparison with Bonferroni correction for multiple comparison, demonstrated that significantly higher levels of DYRK1A were oberved in Dp3Tyb;AppNL-F/NL-F compared to AppNL-F/NL-F cortex (p = 0.002) but that BACE2 levels did not differ between these two genotypes of mice (p = 0.359). There was no effect of an extra copy of the Dp3Tyb region on the abundance of (E) FL-APP level (F(1,49) = 2.183, p = 0.126), (F) CTF-α (F(1,40) = 0.040, p = 0.843), or (G) CTF-β (F(1,40) = 0.008, p = 0.929). As previously reported (Saito, Matsuba et al. 2014), mice homozygous for the AppNL-F allele had lower abundance of (E) FL-APP (F(1,49) = 16.790, p < 0.001), (F) CTF-α (F(1,40) = 15.739, p < 0.001) and a higher abundance of (G) CTF-β (F(1,40) = 147.440, p < 0.001). Wild-type (WT) (female n = 6, male n = 11), Dp3Tyb (female n = 5, male n = 7), AppNL-F (female n = 8, male n = 7) and Dp3Tyb;AppNL-F/NL-F (female n = 5, male n = 8). CTF were below the limit of detection in Wild-type (n = 3) Dp3Tyb (n = 1), AppNL-F (n = 2) and Dp3Tyb;AppNL-F/NL-F (n = 3) samples. Error bars show SEM, data points are independent mice. Dp3Tyb abbreviated to Dp3 for clarity.
Figure 10. BACE2 amyloid-β cleaved fragments do not have increased abundance in the young cortex of the Dp3Tyb;AppNL-F/NL-F model.
LC-MS analysis of immunoprecipated cortical amyloid-β from formic acid fraction normalised to weight of cortical tissue, was used to determine if the Dp3Tyb region was sufficent to alter the abundance of putative BACE2 cleavage products at 3-months of age. (A) Dp3Tyb;AppNL-F/NL-F cortex weighs more than AppNL-F/NL-F cortex at 3-months of age (F(1,22) = 7.772, p = 0.011). AppNL-F/NL-F (female n = 8, male n = 5), Dp3Tyb;AppNL-F/NL-F (female n = 4, male n = 9). No significant increase in the abundance of (B) amyloid-β1-19, (F(1,20) = 0.166 p = 0.688), (C) amyloid-β1-20 (F(1,20) = 0.274 p = 0.607) or (D) amyloid-β1-34 (F(1,20) = 0.005 p = 0.942) was observed. No difference in the abundance of (E) amyloid-β1-4, (F(1,15) = 1.622, p = 0.222), (F) amyloid-β1-15, (F(1,19) = 0.496, p = 0.490), (G) amyloid-β1.16, (F(1,19) = 2.274, p = 0.148) or (H) amyloid-β1.17, (F(1,19) = 0.079, p = 0.781) was detected in the cortex of Dp3Tyb;AppNL-F/NL-F compared with AppNL-F/NL-F mice. AppNL-F/NL-F (female n = 8, male n = 5), Dp3Tyb;AppNL-F/NL-F (female n = 4, male = 9). Dp3Tyb abbreviated to Dp3 for clarity. Error bars show SEM, data points are independent mice. For amyloid-β1-19 and Amyloid-β1-20 n = 1 sample was below the limit of detection per genotype. Amyloid-β1-4 n = 3 samples were below the limit of detection per genotype, amyloid-β1-15, 1-16, 1-17 n = 1 samples were below the limit of detection per genotype. These samples are shown on the graphs but were excluded from ANOVA.
However, we found no evidence of changes to the abundance of humanised-APP or mouse-APP in Dp3Tyb;AppNL-F/NL-F or Dp3Tyb models compared to AppNL-F/NL-F controls in the cortex at 3-months of age (Fig. 7D-E). Similarly no change in human-CTF-β or mouse-CTFβ was observed in the Dp3Tyb model (Fig. 7F, G). We also found no change in human/mouse APP or CTF-β abundance in the Dp(10)2Yey;AppNL-F/NL-F mouse (Fig. 8). We found no evidence of changes to total mouse amyloid-β40 or amyloid-β42 in young Dp3Tyb compared with wild-type controls (3-months of age) or insoluble human amyloid-β40 or amyloid-β42 in young (3-months of age) Dp3Tyb;AppNL-F/NL-F compared with AppNL-F/NL-F controls (Fig. 9, 11). These data suggest that the decreased accumulation of human amyloid-β in the Dp3Tyb model is not likely to be the result of changed abundance of APP, CTF-β or amyloid-β in the young brain and a mechanism that decreases aggregation or enhances clearance of amyloid-β may be causal, consistent with the data from the Tc1;AppNL-F/NL-F model.
Figure 8. The abundance of FL-APP and CTF is not altered by duplication of the Dp10Yey Hsa21 orthologous region.
The abundance of (A, D) full-length, APP (FL-APP), (B, D) C-terminal fragment-α (CTF-α) and (C, D) C-terminal fragment-β (CTF-β) relative to β-actin loading control was measured by western blot in the cortex at 3-months of age in male and female mice. There was no effect of an extra copy of the Dp(10)2Yey region on the abundance of (A) FL-APP level (F(1,22) = 0.828, p = 0.372), (B) CTF-α (F(1,22) = 0.054, p = 0.819) or (C) CTF-β abundance (F(1,22) = 0.829, p = 0.372). As previously observed, mice homozygous for the AppNL-F allele had lower abundance of (A) FL-APP (F(1,22) = 8.168, p = 0.009), (B) CTF-α (F(1,22) = 72.150, p < 0.001) and a higher abundance of (C) CTF-β (F(1,22) = 15.300, p < 0.001). Wild-type (WT) (female n = 1, male n = 4), Dp(10)2Yey (male n = 8), AppNL-F (male n = 9) and Dp(10)2Yey;AppNL-F/NL-F (female n = 2, male n = 4). Error bars show SEM, data points are independent mice. Dp(10)2Yey abbreviated to Dp10 for clarity.
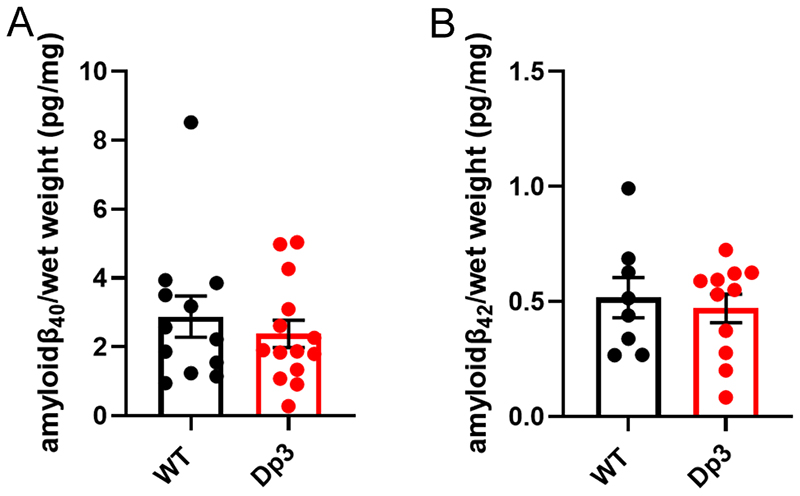
Figure 9. An additional copy of the Dp3Tyb region does not alter the abundance of mouse amyloid-β in the young cortex.
Total cortical proteins were homogenised in TBS buffer, from mice of 3-months of age, and amyloid-β abundance was analysed by MSD assay (4G8). An additional copy of the Dp3Tyb region did not alter the abundance of (A) amyloid-β40 (F1,14) = 0.693, p = 0.419) or (B) amyloid-β42 (F1,7) = 0.410, p = 0.718). Wild-type (WT) (female n = 5, male n = 11), Dp3Tyb (female n = 7, male n = 12). For amyloid-β40 (WT n = 4 and Dp3Tyb = 5) and amyloid-β42 (WT= 8 and Dp3Tyb n = 8) were below the limit of detection. Error bars show SEM, data points are independent mice. Dp3Tyb abbreviated to Dp3 for clarity.
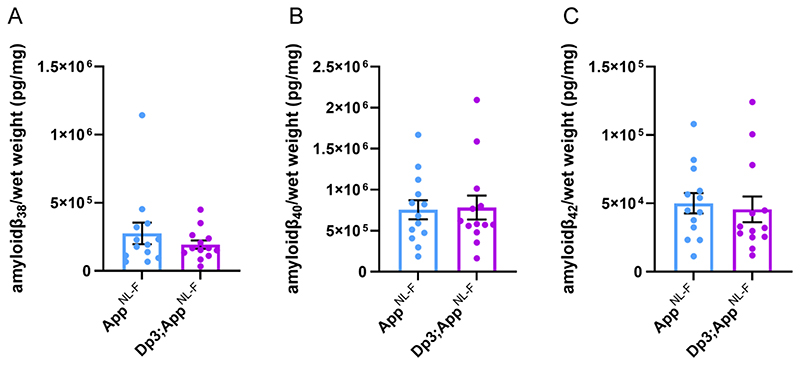
Figure 11. Duplication of the Dp3Tyb region of mouse chromosome 21 orthologous with Hsa21 is not sufficient to decrease amyloid-β biochemical aggregation in the cortex of the AppNL-F/NL-F model at 3-months of age.
Total cortical proteins of 3-month-old mice were prepared for mass-spectrometry analysis and amyloid-β abundance analysed in the formic acid soluble fraction by MSD assay (6E10). No difference in the abundance of formic acid soluble (A) amyloid-β38 (F(1,21) = 1.001, p = 0.328), (B) amyloid-β40 (F(1,21) = 0.032 p = 0.860) or (C) amyloid-β42 (F(1,21) = 0.306, p = 0.443) was detected. AppNL-F/NL-F (female n = 8, male n = 5), Dp3Tyb;AppNL-F/NL-F (female n = 4, male n = 9). Dp3Tyb abbreviated to Dp3 for clarity. Error bars show SEM, data points are independent mice.
In Dp3Tyb mice three-copies of Bace2 do not raise BACE2 abundance or decrease amyloid-β abundance in the young adult brain
The Dp3Tyb model carries an extra copy of Bace2, which encodes a secretase that has been previously reported to cleave APP at the θ site, resulting in the production of amyloid-β1-19 (Sun, He et al. 2006, Alic, Goh et al. 2020). BACE2 has also been suggested to clear amyloid-β, leading to reduced accumulation and production of amyloid-β1-20 and amyloid-β1-34 in an organoid model of DS (Sun, He et al. 2006, Alic, Goh et al. 2020). Thus, we wanted to investigate BACE2 in our AD-DS model. Using western blotting we found that an extra copy of the Dp3Tyb region did not cause BACE2 abundance to be significantly higher in the Dp3Tyb;AppNL-F/NL-F compared with AppNL-F/NL-F cortex (Fig 7A, C), likely because of the underlying high variability in the abundance of this protein in the cortex. We went on to determine if the putative BACE2 amyloid-β degradation products, human-amyloid-β1-20 and human-amyloid-β1-34 or the APP-θ cleavage product human-amyloid-β1-19 were altered by the Dp3Tyb region. No difference in these analytes and human-amyloid-β1-14, human-amyloid-β1-15, human-amyloid-β1-16 or human-amyloid-β1-17 was observed between Dp3Tyb;AppNL-F/NL-F and AppNL-F/NL-F cortex at 3-months of age (Fig. 10).
As we had observed significant variability in the abundance of BACE2 in the cortex, we investigated whether BACE2 protein levels within individual mice predicted the abundance of amyloid-β1-19, amyloid-β1-20 and amyloid-β1-34 in the same animals. No relationships between the level of BACE2 protein and the analytes were observed (amyloid-β1-19 R2 = 0.0001, amyloid-β1-20 R2 = 0.0079, amyloid-β1-34 R2 = 0.0004). These data suggest that differences in BACE2 protein abundance in the young adult mouse brain are not sufficient to cause a detectable alteration in the clearance of amyloid-β or enhanced θ-cleavage. One caveat is that at 3-months of age we cannot yet detect an alteration of the aggregation of human amyloid-β in the Dp3Tyb;AppNL-F/NL-F, as detected by MSD assay after biochemical isolation in formic acid (Fig. 11). Thus, our data could be consistent with a BACE2 aging-dependent mechanism leading to the observed decrease in amyloid-β in the Dp3Tyb;AppNL-F/NL-F model at 8-months of age, but further analysis is required to support this.
Discussion
Here we use a series of mouse models of DS to identify which combination of Hsa21 genes (other than APP) are sufficient to modulate APP/amyloid-β. Our systematic approach identified a region of Hsa21 that contains 38 genes, which is sufficient to decrease the deposition of amyloid-β in vivo at an early time-point. This region contains two lead candidate genes Bace2 and Dyrk1A.
DYRK1A is widely expressed in both the developing and adult mouse brain (Marti, Altafaj et al. 2003, Hammerle, Elizalde et al. 2008) and is found in all major cell types in both the human and mouse brain (Zhang, Chen et al. 2014, Zhang, Sloan et al. 2016). It is a primer kinase that can phosphorylate a large number of proteins, including APP, which has been suggested to increase the protein’s abundance (Ryoo, Cho et al. 2008, Branca, Shaw et al. 2017). Inhibition of the kinase in a transgenic mouse model of AD decreased amyloid-β accumulation (Branca, Shaw et al. 2017, Velazquez, Meechoovet et al. 2019). Consistent with this, a decrease in APP and amyloid-β abundance is observed in a mouse model of DS with a normalised Dyrk1a gene dose (Garcia-Cerro, Rueda et al. 2017). In the Dp3Tyb model, which contains an additional copy of Dyrk1a, we observe no increase in APP abundance (endogenous mouse APP and partially humanised APPSW) or evidence of enhanced amyloid-β accumulation. This suggests that in our knock-in model system, 3 copies of Dyrk1a are not sufficient to modulate APP protein abundance or promote amyloid-β accumulation. Notably, inhibition of DYRK1A has been suggested as both a possible therapeutic to enhance cognition in people who have DS and as a possible AD therapeutic strategy (De la Torre, De Sola et al. 2014, Yin, Jin et al. 2017, Neumann, Gourdain et al. 2018, Nguyen, Duchon et al. 2018). Our data do not support the use of DYRK1A inhibitors as a treatment strategy to decrease early amyloid-β accumulation in people who have DS. However, inhibiting the kinase may have other beneficial effects, such as to slow or prevent the formation of tau neurofibrillary tangles or to alter the response of cells within the brain to accumulating pathology. Further research is needed, in next generation AD-DS mouse models, to address these key questions.
BACE2 has been reported to be expressed in a subset of astrocytes and neurons in the human brain (Holler, Webb et al. 2012). In the mouse the gene is expressed in a subset of neurons with the highest level in the CA3 and subiculum and some expression is also reported in oligodendrocytes and astrocytes lining the lateral ventricles (Voytyuk, Mueller et al. 2018). The protein may function as a θ-secretase (decreasing CTF-β), a β-secretase (increasing CTF-β), and as an amyloid-β degrading protease (decreasing amyloid-β) (Sun, He et al. 2006, Abdul-Hay, Sahara et al. 2012); which mechanism predominates in the human brain is not well understood. A previous study in which BACE2 was overexpressed in a wild-type APP over-expressing mouse model reported no evidence of altered amyloid-β40 or amyloid-β42 abundance in the brain (Azkona, Levannon et al. 2010). Conversely, knocking-down Bace2 in the AppNL-G-F mouse model of amyloid-β accumulation has been reported to decrease CTF-β and soluble amyloid-β (Wang, Xu et al. 2019). In contrast, reducing BACE2 copy number in human organoids produced from individuals with DS or APP-duplication increased amyloid-β and triggered the formation of deposits within the model system. The authors propose this occurs because of the amyloid clearance function of BACE2, as evidenced by the raised levels of amyloid-β degradation products in trisomic compared to isogenic disomic control (Alic, Goh et al. 2020).
In the Dp3Tyb model that contains an additional copy of Bace2, we observe no significant change in CTF-β (endogenous mouse APP and partially humanised APPSW), soluble amyloid-β (endogenous mouse and partially humanised APPSW) or amyloid-β degradation products in the young adult brain. However, we observe a significant decrease in amyloid-β deposition in older mice (8-months of age) consistent with the role of BACE2 as an amyloid-β degrading protease (Alic, Goh et al. 2020). Further studies are required in aged mice to determine if enhanced amyloid-clearance can be detected, or if BACE2 gene-dose correction is sufficient to reverse the reduction in amyloid accumulation caused by the Dp3Tyb model.
Genes other than Dyrk1a or Bace2 may be responsible for the decreased amyloid-β accumulation observed in our model systems, perhaps via enhancing amyloid-β clearance pathways. However, we previously studied the rate of extracellular clearance of human amyloid-β in an alternative model system (cross of the Tc1 with the J20 APP transgenic model) and found no evidence that the amyloid-β clearance rate was modulated by the extra copy of Hsa21 genes (Wiseman, Pulford et al. 2018). In addition, in the same model system we found no evidence of increased abundance of the key clearance enzymes neprilysin or insulin degrading enzyme in the young adult brain. Changes to intracellular clearance/formation of amyloid-β may contribute to the alterations in accumulation of the peptide reported here. Notably DS is associated with significant alterations to endosomal biology (Botte and Potier 2020); neuronal endosomes have a key role in amyloid-β formation and astrocyte and microglia endosomes in amyloid-β clearance. Further studies focussing on cell type specific effects of extra chromosome 21 genes are required to understand this complex and important biology.
DS is caused by the increase in the abundance of Hsa21 gene products; however, not all genes on the chromosome are dosage-sensitive in all contexts. For example, APP dosage sensitivity has been reported to vary over life-span (Choi, Berger et al. 2009) and BACE2 has been reported to be both dosage-sensitive (Barbiero, Benussi et al. 2003) and insensitive in the human brain (Cheon, Dierssen et al. 2008, Holler, Webb et al. 2012). In contrast, the abundance of DYRK1A is highly sensitive to gene dose in the vast majority of reported studies (Arbones, Thomazeau et al. 2019), including previously in the adult brain of the Tc1 mouse model (Sheppard, Plattner et al. 2012, Ahmed, Dhanasekaran et al. 2013, Naert, Ferre et al. 2018). Consistent with this, here we report a significant increase in DYRK1A in the adult brain of the new Dp3Tyb model. However we could not detect a significant increase in BACE2, this lack of statistical significance is likely caused by the higher intra-animal variation in abundance of this protein in the mouse brain. Similar overlapping levels of Bace2 gene expression have previously been reported at RNA level in the brain of an alternative mouse model of DS compared with euploid controls (Sultan, Piccini et al. 2007).
The relative abundance of proteins in AD-DS model systems is particularly important, as Down syndrome is caused by an inbalance in the relative abundance of gene products. Previous research using APP transgenic models, which over express APP protein, may mask the subtle but physiologically relevant effects of the 50% increase in the abundance of other Hsa21 gene products. This study addresses this limitation, here we systematically investigate the effect of additional copies of Hsa21 gene orthologues, other than App, on APP biology in a knock-in mouse model system. However, because of technical limitations we were not able to study the effect of an extra copy of genes located close to App, including the role of key genes such as Adamts1 (Kunkle, Grenier-Boley et al. 2019) and Usp25 (Zheng, Li et al. 2021). We were also unable to determine if genes located far apart on the chromosome act synergistically to cause a modulation of APP biology. Thus our data does not preclude a multigenic role for the genes in the Dp(10)2Yey or Dp(17)3Yey regions when combined with other Hsa21 genes. A further limitation is the use of AD-associated mutations in APP to drive pathology as these mutations alter the subcellular localisation and processing of APP (Sasaguri, Nilsson et al. 2017), which may modulate the effect of trisomy of Hsa21 on these processes. We have partially addressed this issue by undertaking a side-by-side comparison of mouse APP/amyloid-β and partially humanised APP/amyloid-β.
We studied two time-points in our model systems. Firstly, the young adult brain at 3 months of age, prior to the accumulation of human amyloid-β in the AppNL-F in order to understand the modulation of early upstream processes by the additional copies of Hsa21 orthologues. Secondly the middle-aged adult brain at 8-months of age, which in the AppNL-F mouse, models the initiation of amyloid-β accumulation (Saito, Matsuba et al. 2014). Thus in this study we focus on how additional copies of Hsa21 gene orthologues modulate the earliest stages of AD pathology; the formation and early accumulation of amyloid-β in the brain. We note that the effect of the additional genes may differ later in disease, including during the maturation of amyloid-β plaques, the formation of tau neurofibrillary tangles and the response of astrocytes, microglia and neurons to AD pathology. We studied both male and female mice, to ensure the generalisability of our findings, as sex is know to modulate APP/amyloid-β in mouse models (Jankowsky and Zheng 2017). We did not find evidence that sex modulated the effect of additional Hsa21 genes on amyloid accumulation in either the Tc1 or Dp3 models.
Our results indicate that an additional copy of genes on Hsa21 in people who have DS can modulate key AD biology. The extent of this effect is likely to differ between individuals both because of genetic variation on Hsa21 and differences in life-style factors that modulate dementia risk. This may contribute to the variation in age of onset of both pathology and dementia observed in people who have DS.
Conclusion
DS is a complex condition that alters multiple aspects of neurobiology and physiology. Here we demonstrate using physiological mouse models that an additional copy of Hsa21 genes reduces the accumulation of amyloid-β within the brain; one of the earliest steps in the AD pathogenic process. Thus trisomy of Hsa21 may partially protect individuals who have DS from the accumulation of amyloid-β, resultant from their extra copy of APP. Moreover we predict that individuals who have DS will accumulate amyloid-β more slowly than other groups who develop other genetic forms of early-onset AD, including that caused by duplication of APP. However, multiple studies of adults who have DS demonstrate that these individuals do accumulate substantial amyloid-β within their brains by mid-life, thus the effect of an additional copy of other genes on the chromosome is not sufficient to delay pathology development and AD primary prevention therapies targeting amyloid-β are likely to significantly benefit individuals who have DS (Fortea, Zaman et al. 2021). Treatments for AD-DS and other medical conditions associated with DS must take into account the differences in biology of individuals with the syndrome; to ensure that interventions do not reverse the beneficial effect of the additional genes.
Significance Statement.
Trisomy of chromosome 21 is a commonly occurring genetic risk factor for early-onset Alzheimer’s disease, which has been previously attributed to people with Down syndrome having three copies of the APP gene, which is encoded on chromosome 21. However, we have shown that an extra copy of other chromosome 21 genes modifies AD-like phenotypes independently of APP copy number (Wiseman et al. 2018, Brain; Tosh et al. 2021 Scientific Reports). Here, we use a mapping approach to narrow-down the genetic cause of the modulation of pathology; demonstrating that gene(s) on chromosome 21 decrease amyloid-β accumulation in the brain, independently of alterations to full-length APP or C-terminal fragment abundance and that just 38 genes are sufficient to cause this.
Table 2. Summary of negative control samples, which do not carry an AppNL-F allele and do not produce human amyloid-β, used in MSD assays.
Negative control samples from mice that do not carry the AppNL-F allele and do not produce human amyloid-β were assayed by 6E10 MSD assay alongside samples homozygous for the AppNL-F allele and which do produce human amyloid-β. The epitope for 6E10 lies within the region of sequence difference between human and mouse Aβ, the antibody is against the human sequence. In some cases as indicated all samples without an AppNL-F allele were below the limit of detection for the assay and thus mean and standard deviations are not available (N/A)
| Figure | Measurement | Genotype | Total ‘n’ | Below the limit of detection | Above Limit of Detection | ||
|---|---|---|---|---|---|---|---|
| n | Mean (pg/mg) | Standard deviation | |||||
| Figure 5A | Gnd HCl amyloid-β42/ total brain weight | WT | 4 | 4 | 0 | N/A | N/A |
| Figure 5A | Gnd HCl amyloid-β42/ total brain weight | Dp3Tyb | N/A | N/A | N/A | N/A | N/A |
| Figure 5B | Gnd HCl amyloid-β42/total brain weight | WT | 6 | 0 | 6 | 0.505 | 0.444 |
| Figure 5B | Gnd HCl amyloid-β42/ total brain weight | Dp(10)2Tyb | 3 | 0 | 3 | 0.59 | 0.411 |
| Figure 5C | Gnd HCl amyloid-β42/ total brain weight | WT | 5 | 4 | 1 | 6.154 | N/A |
| Figure 5C | Gnd HCl amyloid-β42/ total brain weight | Dp(17)3Tyb | 5 | 5 | 0 | N/A | N/A |
| Figure 5D | 1% Triton amyloid-β42/total brain weight | WT | 4 | 3 | 1 | 0.002 | N/A |
| Figure 5D | 1% Triton amyloid-β42/ total brain weight | Dp3Tyb | Not available | N/A | N/A | N/A | N/A |
| Figure 5E | 1% Triton amyloid-β42/ total brain weight | WT | 6 | 5 | 1 | 0.009 | N/A |
| Figure 5E | 1% Triton amyloid-β42/ total brain weight | Dp(10)2Tyb | 3 | 2 | 1 | 0.007 | N/A |
| Figure 5F | 1% Triton amyloid-β42/ total brain weight | WT | 5 | 2 | 3 | 0.475 | 0.814 |
| Figure 5F | 1% Triton amyloid-β42/ total brain weight | Dp(17)3Tyb | 5 | 3 | 2 | 0.074 | N/A |
| Figure 5G | Tris amyloid-β42/ total brain weight | WT | 4 | 3 | 1 | 0.015 | N/A |
| Figure 5G | Tris amyloid-β42/ total brain weight | Dp3Tyb | Not available | N/A | N/A | N/A | N/A |
| Figure 5H | Tris amyloid-β42/total brain weight | WT | 6 | 5 | 1 | 0.006 | N/A |
| Figure 5H | Tris amyloid-β42/ total brain weight | Dp(10)2Tyb | 3 | 2 | 1 | 0.007 | N/A |
| Figure 5I | Tris amyloid-β42/ total brain weight | WT | 5 | 2 | 3 | 0.304 | 0.517 |
| Figure 5I | Tris amyloid-β42/ total brain weight | Dp(17)3Tyb | 5 | 2 | 3 | 0.011 | 0.009 |
| Figure 6A | Gnd HCl amyloid-β40/total brain weight | WT | 6 | 0 | 6 | 2.097 | 1.246 |
| Figure 6A | Gnd HCl amyloid-β40/total brain weight | Dp(10)2Tyb | 3 | 0 | 3 | 2.255 | 1.796 |
| Figure 6B | Gnd HCl amyloid-β40/ total brain weight | WT | 5 | 5 | 0 | N/A | N/A |
| Figure 6B | Gnd HCl amyloid-β40/ total brain weight | Dp(17)3Tyb | 5 | 5 | 0 | N/A | N/A |
| Figure 6C | 1% Triton amyloid-β42/ total brain weight | WT | 6 | 4 | 2 | 0.058 | 0.004 |
| Figure 6C | 1% Triton amyloid-β42/ total brain weight | Dp(10)2Tyb | 3 | 1 | 2 | 0.035 | 0.023 |
| Figure 6D | 1% Triton amyloid-β42/ total brain weight | WT | 5 | 4 | 1 | 3.241 | N/A |
| Figure 6D | 1% Triton amyloid-β42/ total brain weight | Dp(17)3Tyb | 5 | 5 | 5 | N/A | N/A |
Abbreviations
- AD
Alzheimer’s disease
- AD-DS
Alzheimer’s disease in Down syndrome
- APP
Amyloid precursor protein
- BSA
Bovine serum albumin
- DS
Down syndrome
- FA
Formic acid
- IP
Immunoprecipitation
- LC-MS
Liquid chromatography-mass spectrometry
- MS
Mass spectrometry
- MSD
Meso Scale Discovery
- TBS
Phosphate-buffered saline (PBS) Standard error of the mean (SEM) Tris-buffered saline
Footnotes
Competing interests:
H.Z. has served at scientific advisory boards and/or as a consultant for Abbvie, Alector, Annexon, Artery Therapeutics, AZTherapies, CogRx, Denali, Eisai, Nervgen, Pinteon Therapeutics, Red Abbey Labs, Passage Bio, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure and Biogen, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). KB has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Prothena, Roche Diagnostics, and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program, all unrelated to the work presented in this paper.
Author Contributions
P.M. undertook biochemical and histological experiments, data analysis and edited the manuscript. J.T and S.A undertook biochemical and histological experiments. G.L undertook histological analysis. S.N undertook histological experiments and K.C. undertook genotyping and edited the manuscript. E.G.W. and G.B. performed the LC-MS analysis. E.Y., T. S. and T.C.S. contributed essential research resources, E.M.C.F. and V.L.J.T, contributed essential research resources, designed and supervised the study and edited the manuscript. F.K.W. designed and supervised the study, undertook data analysis and wrote the manuscript. All authors revised the manuscript.
Acknowledgments
F.K.W., H.Z. and P.M. are supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK (UKDRI-1014) and by an Alzheimer’s Research UK Senior Research Fellowship (ARUK-SRF2018A-001). F.K.W. also received funding that contributed to the work in this paper from the MRC via CoEN award MR/S005145/1. J.L.T. was funded by an Alzheimer’s Society Ph.D. studentship awarded to F.K.W. and E.M.C.F. The authors were funded by a Wellcome Trust Strategic Award (grant number: 098330/Z/12/Z) awarded to The London Down Syndrome (LonDownS) Consortium (V.L.J.T., and E.M.C.F). Additionally, the authors were funded by a Wellcome Trust Joint Senior Investigators Award (V.L.J.T. and E.M.C.F., grant numbers: 098328, 098327), the Medical Research Council (programme number U117527252; awarded to V.L.J.T). V.L.J.T. is also funded by the Francis Crick Institute which receives its core funding from the Medical Research Council (FC001194), Cancer Research UK (FC001194) and the Wellcome Trust (FC001194). H.Z. is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the AD Strategic Fund and the Alzheimer’s Association (#ADSF-21-831376-C, #ADSF-21-831381-C and #ADSF-21-831377-C), the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019-0228), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE), and the UK Dementia Research Institute at UCL. KB is supported by the Swedish Research Council (#2017-00915), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), and the Alzheimer’s Association 2021 Zenith Award (ZEN-21-848495).
For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript version arising from this submission. All authors read and approved the final manuscript. We thank Dr. Amanda Heslegrave (UCL) for assistance with this project.
References
- Abdul-Hay SO, Sahara T, McBride M, Kang D, Leissring MA. Identification of BACE2 as an avid ss-amyloid-degrading protease. Mol Neurodegener. 2012;7:46. doi: 10.1186/1750-1326-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed MM, Dhanasekaran AR, Tong S, Wiseman FK, Fisher EM, Tybulewicz VL, Gardiner KJ. Protein profiles in Tc1 mice implicate novel pathway perturbations in the Down syndrome brain. Hum Mol Genet. 2013;22(9):1709–1724. doi: 10.1093/hmg/ddt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic I, Goh PA, Murray A, Portelius E, Gkanatsiou E, Gough G, Mok KY, Koschut D, Brunmeir R, Yeap YJ, et al. Patient-specific Alzheimer-like pathology in trisomy 21 cerebral organoids reveals BACE2 as a gene dose-sensitive AD suppressor in human brain. Mol Psychiatry. 2020 doi: 10.1038/s41380-020-0806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbones ML, Thomazeau A, Nakano-Kobayashi A, Hagiwara M, Delabar JM. DYRK1A and cognition: A lifelong relationship. Pharmacol Ther. 2019;194:199–221. doi: 10.1016/j.pharmthera.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Azkona G, Levannon D, Groner Y, Dierssen M. In vivo effects of APP are not exacerbated by BACE2 co-overexpression: behavioural characterization of a double transgenic mouse model. Amino Acids. 2010;39(5):1571–1580. doi: 10.1007/s00726-010-0662-8. [DOI] [PubMed] [Google Scholar]
- Barbiero L, Benussi L, Ghidoni R, Alberici A, Russo C, Schettini G, Pagano SF, Parati EA, Mazzoli F, Nicosia F, Signorini S, et al. BACE-2 is overexpressed in Down’s syndrome. Exp Neurol. 2003;182(2):335–345. doi: 10.1016/s0014-4886(03)00049-9. [DOI] [PubMed] [Google Scholar]
- Benejam B, Videla L, Vilaplana E, Barroeta I, Carmona-Iragui M, Altuna M, Valldeneu S, Fernandez S, Gimenez S, Iulita F, Garzon D, et al. Diagnosis of prodromal and Alzheimer’s disease dementia in adults with Down syndrome using neuropsychological tests. Alzheimers Dement (Amst) 2020;12(1):e12047. doi: 10.1002/dad2.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botte A, Potier MC. Focusing on cellular biomarkers: The endo-lysosomal pathway in Down syndrome. Prog Brain Res. 2020;251:209–243. doi: 10.1016/bs.pbr.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Branca C, Shaw DM, Belfiore R, Gokhale V, Shaw AY, Foley C, Smith B, Hulme C, Dunckley T, Meechoovet B, Caccamo A, et al. Dyrk1 inhibition improves Alzheimer’s disease-like pathology. Aging Cell. 2017;16(5):1146–1154. doi: 10.1111/acel.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon MS, Dierssen M, Kim SH, Lubec G. Protein expression of BACE1, BACE2 and APP in Down syndrome brains. Amino Acids. 2008;35(2):339–343. doi: 10.1007/s00726-007-0618-9. [DOI] [PubMed] [Google Scholar]
- Choi JH, Berger JD, Mazzella MJ, Morales-Corraliza J, Cataldo AM, Nixon RA, Ginsberg SD, Levy E, Mathews PM. Age-dependent dysregulation of brain amyloid precursor protein in the Ts65Dn Down syndrome mouse model. J Neurochem. 2009;110(6):1818–1827. doi: 10.1111/j.1471-4159.2009.06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson YS, Robinson A, Prasher VP, Mann DMA. The age of onset and evolution of Braak tangle stage and Thal amyloid pathology of Alzheimer’s disease in individuals with Down syndrome. Acta Neuropathol Commun. 2018;6(1):56. doi: 10.1186/s40478-018-0559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf G, Buckley F, Skotko BG. Estimation of the number of people with Down syndrome in Europe. Eur J Hum Genet. 2021;29(3):402–410. doi: 10.1038/s41431-020-00748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Torre R, De Sola S, Pons M, Duchon A, de Lagran MM, Farre M, Fito M, Benejam B, Langohr K, Rodriguez J, Pujadas M, et al. Epigallocatechin-3-gallate, a DYRK1A inhibitor, rescues cognitive deficits in Down syndrome mouse models and in humans. Mol Nutr Food Res. 2014;58(2):278–288. doi: 10.1002/mnfr.201300325. [DOI] [PubMed] [Google Scholar]
- Doran E, Keator D, Head E, Phelan MJ, Kim R, Totoiu M, Barrio JR, Small GW, Potkin SG, Lott IT. Down Syndrome, Partial Trisomy 21, and Absence of Alzheimer’s Disease: The Role of APP. J Alzheimers Dis. 2017;56(2):459–470. doi: 10.3233/JAD-160836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortea J, Zaman SH, Hartley S, Rafii MS, Head E, Carmona-Iragui M. Alzheimer’s disease associated with Down syndrome: a genetic form of dementia. Lancet Neurol. 2021;20(11):930–942. doi: 10.1016/S1474-4422(21)00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cerro S, Rueda N, Vidal V, Lantigua S, Martinez-Cue C. Normalizing the gene dosage of Dyrk1A in a mouse model of Down syndrome rescues several Alzheimer’s disease phenotypes. Neurobiol Dis. 2017;106:76–88. doi: 10.1016/j.nbd.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Gkanatsiou E, Portelius E, Toomey CE, Blennow K, Zetterberg H, Lashley T, Brinkmalm G. A distinct brain beta amyloid signature in cerebral amyloid angiopathy compared to Alzheimer’s disease. Neurosci Lett. 2019;701:125–131. doi: 10.1016/j.neulet.2019.02.033. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer’s disease and Down’s syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122(3):1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- Gribble SM, Wiseman FK, Clayton S, Prigmore E, Langley E, Yang F, Maguire S, Fu B, Rajan D, Sheppard O, Scott C, et al. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One. 2013;8(4):e60482. doi: 10.1371/journal.pone.0060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj F, Pereira PL, Najas S, Barallobre MJ, Chabert C, Souchet B, Sebrie C, Verney C, Herault Y, Arbones M, Delabar JM. DYRK1A: a master regulatory protein controlling brain growth. Neurobiol Dis. 2012;46(1):190–203. doi: 10.1016/j.nbd.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Hammerle B, Elizalde C, Tejedor FJ. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci. 2008;27(5):1061–1074. doi: 10.1111/j.1460-9568.2008.06092.x. [DOI] [PubMed] [Google Scholar]
- Holler CJ, Webb RL, Laux AL, Beckett TL, Niedowicz DM, Ahmed RR, Liu Y, Simmons CR, Dowling AL, Spinelli A, Khurgel M, et al. BACE2 expression increases in human neurodegenerative disease. Am J Pathol. 2012;180(1):337–350. doi: 10.1016/j.ajpath.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtta M, Hansson O, Andreasson U, Hertze J, Minthon L, Nagga K, Andreasen N, Zetterberg H, Blennow K. Evaluating amyloid-beta oligomers in cerebrospinal fluid as a biomarker for Alzheimer’s disease. PLoS One. 2013;8(6):e66381. doi: 10.1371/journal.pone.0066381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol Neurodegener. 2017;12(1):89. doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana-Elola E, Watson-Scales S, Slender A, Gibbins D, Martineau A, Douglas C, Mohun T, Fisher EM, Tybulewicz V. Genetic dissection of Down syndrome-associated congenital heart defects using a new mouse mapping panel. Elife. 2016;5:e11614. doi: 10.7554/eLife.11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E, Altafaj X, Dierssen M, de la Luna S, Fotaki V, Alvarez M, Perez-Riba M, Ferrer I, Estivill X. Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 2003;964(2):250–263. doi: 10.1016/s0006-8993(02)04069-6. [DOI] [PubMed] [Google Scholar]
- McCarron M, McCallion P, Reilly E, Dunne P, Carroll R, Mulryan N. A prospective 20-year longitudinal follow-up of dementia in persons with Down syndrome. J Intellect Disabil Res. 2017;61(9):843–852. doi: 10.1111/jir.12390. [DOI] [PubMed] [Google Scholar]
- Naert G, Ferre V, Keller E, Slender A, Gibbins D, Fisher EM, Tybulewicz VL, Maurice T. In vivo and ex vivo analyses of amyloid toxicity in the Tc1 mouse model of Down syndrome. J Psychopharmacol. 2018;32(2):174–190. doi: 10.1177/0269881117743484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F, Gourdain S, Albac C, Dekker AD, Bui LC, Dairou J, Schmitz-Afonso I, Hue N, Rodrigues-Lima F, Delabar JM, Potier MC, et al. DYRK1A inhibition and cognitive rescue in a Down syndrome mouse model are induced by new fluoro-DANDY derivatives. Sci Rep. 2018;8(1):2859. doi: 10.1038/s41598-018-20984-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TL, Duchon A, Manousopoulou A, Loaec N, Villiers B, Pani G, Karatas M, Mechling AE, Harsan LA, Limanton E, Bazureau JP, et al. Correction of cognitive deficits in mouse models of Down syndrome by a pharmacological inhibitor of DYRK1A. Dis Model Mech. 2018;11(9) doi: 10.1242/dmm.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, Cooke S, Sesay A, Modino S, Vanes L, Hernandez D, Linehan JM, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309(5743):2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18(7):e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E, Tran AJ, Andreasson U, Persson R, Brinkmalm G, Zetterberg H, Blennow K, Westman-Brinkmalm A. Characterization of amyloid beta peptides in cerebrospinal fluid by an automated immunoprecipitation procedure followed by mass spectrometry. J Proteome Res. 2007;6(11):4433–4439. doi: 10.1021/pr0703627. [DOI] [PubMed] [Google Scholar]
- Ryoo SR, Cho HJ, Lee HW, Jeong HK, Radnaabazar C, Kim YS, Kim MJ, Son MY, Seo H, Chung SH, Song WJ. Dual-specificity tyrosine(Y)-phosphorylation regulated kinase 1A-mediated phosphorylation of amyloid precursor protein: evidence for a functional link between Down syndrome and Alzheimer’s disease. J Neurochem. 2008;104(5):1333–1344. doi: 10.1111/j.1471-4159.2007.05075.x. [DOI] [PubMed] [Google Scholar]
- Saito T, Matsuba Y, Mihira N, Takano J, Nilsson P, Itohara S, Iwata N, Saido TC. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17(5):661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- Sasaguri H, Nilsson P, Hashimoto S, Nagata K, Saito T, De Strooper B, Hardy J, Vassar R, Winblad B, Saido TC. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017;36(17):2473–2487. doi: 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serneels L, T’Syen D, Perez-Benito L, Theys T, Holt MG, De Strooper B. Modeling the beta-secretase cleavage site and humanizing amyloid-beta precursor protein in rat and mouse to study Alzheimer’s disease. Mol Neurodegener. 2020;15(1):60. doi: 10.1186/s13024-020-00399-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard O, Plattner F, Rubin A, Slender A, Linehan JM, Brandner S, Tybulewicz VL, Fisher EM, Wiseman FK. Altered regulation of tau phosphorylation in a mouse model of down syndrome aging. Neurobiol Aging. 2012;33(4):828 e831-844. doi: 10.1016/j.neurobiolaging.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Piccini I, Balzereit D, Herwig R, Saran NG, Lehrach H, Reeves RH, Yaspo ML. Gene expression variation in Down’s syndrome mice allows prioritization of candidate genes. Genome Biol. 2007;8(5):R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, He G, Song W. BACE2, as a novel APP theta-secretase, is not responsible for the pathogenesis of Alzheimer’s disease in Down syndrome. FASEB J. 2006;20(9):1369–1376. doi: 10.1096/fj.05-5632com. [DOI] [PubMed] [Google Scholar]
- Tosh JL, Rhymes ER, Mumford P, Whittaker HT, Pulford LJ, Noy SJ, Cleverley K, Walker MC, Tybulewicz VLJ, Wykes RC, Fisher EMC, et al. Genetic dissection of down syndrome-associated alterations in APP/amyloid-beta biology using mouse models. Sci Rep. 2021;11(1):5736. doi: 10.1038/s41598-021-85062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez R, Meechoovet B, Ow A, Foley C, Shaw A, Smith B, Oddo S, Hulme C, Dunckley T. Chronic Dyrk1 Inhibition Delays the Onset of AD-Like Pathology in 3xTg-AD Mice. Mol Neurobiol. 2019;56(12):8364–8375. doi: 10.1007/s12035-019-01684-9. [DOI] [PubMed] [Google Scholar]
- Voytyuk I, Mueller SA, Herber J, Snellinx A, Moechars D, van Loo G, Lichtenthaler SF, De Strooper B. BACE2 distribution in major brain cell types and identification of novel substrates. Life Sci Alliance. 2018;1(1):e201800026. doi: 10.26508/lsa.201800026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu Q, Cai F, Liu X, Wu Y, Song W. BACE2, a conditional beta-secretase, contributes to Alzheimer’s disease pathogenesis. JCI Insight. 2019;4(1) doi: 10.1172/jci.insight.123431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman FK, Al-Janabi T, Hardy J, Karmiloff-Smith A, Nizetic D, Tybulewicz VL, Fisher EM, Strydom A. A genetic cause of Alzheimer disease: mechanistic insights from Down syndrome. Nat Rev Neurosci. 2015;16(9):564–574. doi: 10.1038/nrn3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman FK, Pulford LJ, Barkus C, Liao F, Portelius E, Webb R, Chavez-Gutierrez L, Cleverley K, Noy S, Sheppard O, Collins T, et al. Trisomy of human chromosome 21 enhances amyloid-beta deposition independently of an extra copy of APP. Brain. 2018;141(8):2457–2474. doi: 10.1093/brain/awy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Jin N, Shi J, Zhang Y, Wu Y, Gong CX, Iqbal K, Liu F. Dyrk1A overexpression leads to increase of 3R-tau expression and cognitive deficits in Ts65Dn Down syndrome mice. Sci Rep. 2017;7(1):619. doi: 10.1038/s41598-017-00682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Li Z, Jia Z, Clapcote SJ, Liu C, Li S, Asrar S, Pao A, Chen R, Fan N, Carattini-Rivera S, et al. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19(14):2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA, 3rd, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89(1):37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Li G, Wang S, Zhou Y, Liu K, Gao Y, Zhou Y, Zheng L, Zhu L, Deng Q, Wu M, et al. Trisomy 21-induced dysregulation of microglial homeostasis in Alzheimer’s brains is mediated by USP25. Sci Adv. 2021;7(1) doi: 10.1126/sciadv.abe1340. [DOI] [PMC free article] [PubMed] [Google Scholar]