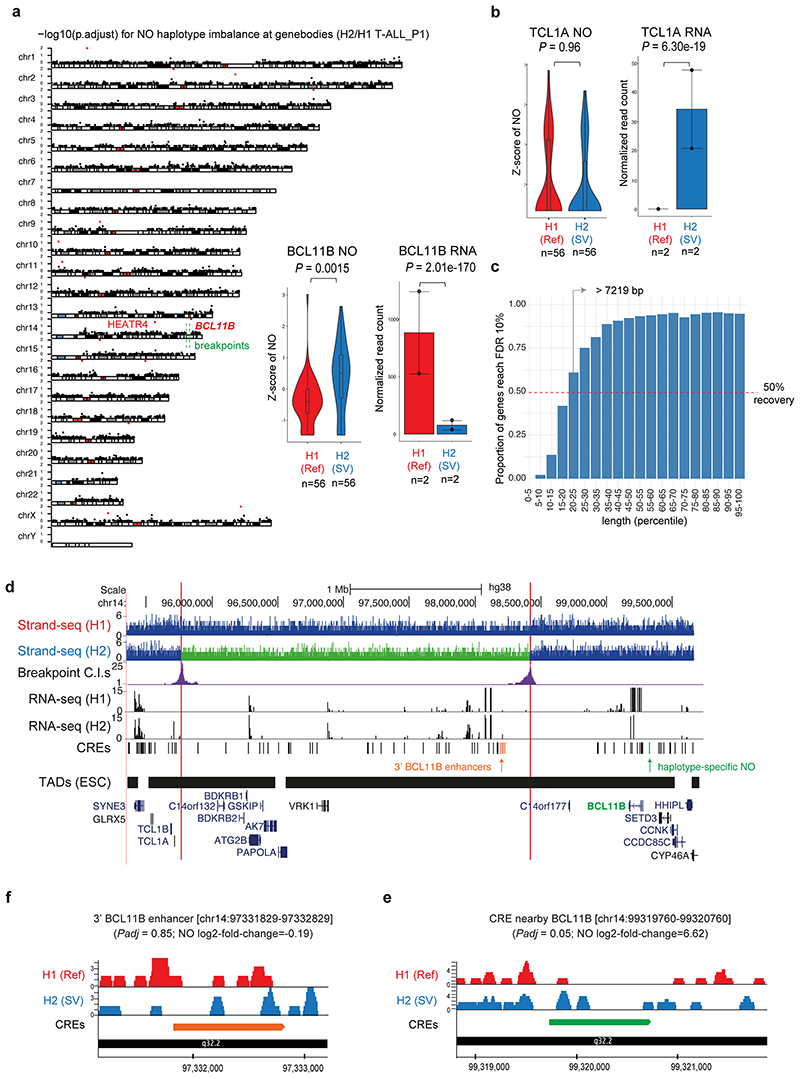
Extended Data Fig. 6. Haplotype-specific NO analysis in T-ALL_P1.
(a) For each chromosomal karyogram, the y-axis indicates the significance of haplotype-specific NO at each gene (-log10 p.adjust). Genes with haplotype-specific NO are indicated using red dots (FDR 10%). An inlet figure depicts haplotype-specific NO (two-sided wilcoxon ranksum test and Benjamini Hochberg multiple correction; n = 56 cells) and RNA expression at the BCL11B gene locus (two-sided likelihood ratio test and Benjamini Hochberg multiple correction; n = 2 biological replicates), which has a nearby somatic SV (within 1 Megabase) and represents the (only) predicted local SV effect. (b) We did not measure haplotype-specific NO for TCL1A (two-sided wilcoxon ranksum test and Benjamini Hochberg multiple correction; n = 56 cells), a small gene with 4229 bp in size, in spite of its haplotype-specific gene expression24 (two-sided likelihood ratio test and Benjamini Hochberg multiple correction; n = 2 biological replicates). Boxplots were defined by minima=25th percentile-1.5X interquartile range (IQR), maxima=75th percentile+1.5X IQR, center=median, and bounds of box=25th and 75th percentile. For bargraphs, data are presented as mean values +/- SEM (a-b). (c) Simulation analysis revealed a minimum gene length (7219 bp) needed to robustly detect haplotype-specific NO at gene bodies, a gene length met by 80% of genes in the genome (Supplementary Notes). (d) Inversion breakpoints and rearranged TADs. Known 3’ BCL11B enhancers96 are depicted in orange. In the not rearranged haplotype, they are located proximal to BCL11B, but in the inverted haplotype these enhancers they are located far away from BCL11B, and proximal to TCL1A in the different TAD boundary. (e) Application of scNOVA identified an intergenic CRE near the BCL11B with haplotype-specific NO. The browser track depicts the haplotype-resolved NO of the not rearranged (Ref) homolog in red and the SV homolog in blue. (f) The known 3’ BCL11B enhancer does not show significant haplotype-specific NO, but the inversion physically relocates these enhancers to the far distance from the BCL11B. A representative CRE is shown amongst four CREs overlapping with known 3’ BCL11B enhancers.