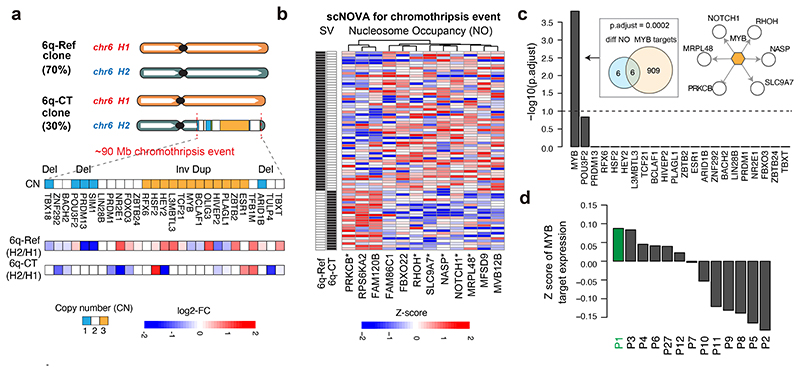
Figure 5. scNOVA identifies functional effects of a subclonal chromothripsis event.
(a) 27 TF genes located in a segment that underwent chromothripsis24 on 6q in T-ALL_P1. Haplotype-specific NO measurements, which scNOVA generated for CREs assigned to the nearest genes, are depicted below. FC: fold-change of normalized haplotype-specific NO (shown for each subclone). 6q-CT: subclone bearing chromothripsis on 6q. 6q-Ref: subclone bearing a not rearranged chromosome 6. (b) Heatmap of 12 genes with differential activity between subclones in T-ALL_P1, based on scNOVA (denoted CT gene signature). Asterisks denote TF targets highlighted in (c). (c) TF target over-representation analyses for CT gene signature, revealing c-Myb as the only significant hit. Venn diagram depicts enrichment of c-Myb targets (P-value based on an FDR-adjusted hypergeometric test). Upper right: network with c-Myb and its target genes based on scNOVA, combined with prior knowledge. (d) Mean Z-scores of c-Myb target gene expression measured by bulk RNA-seq in a panel of 13 T-ALL-derived samples. T-ALL_P1 (P1) exhibited the overall highest expression of c-Myb targets.