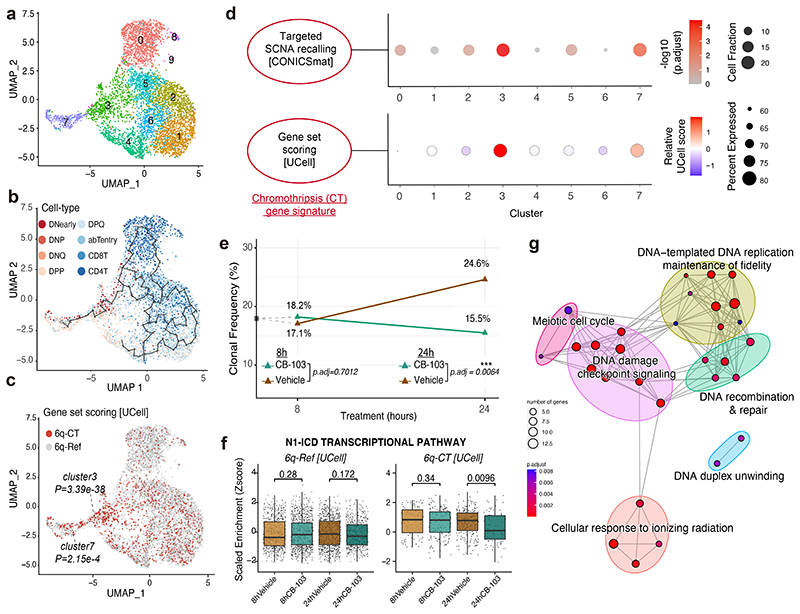
Figure 6. Targeting the chromothriptic subclone in cell culture.
(a) UMAP of scRNA-seq data showing ten unsupervised clusters in T-ALL_P1. (b) Overlay of gene set-derived cell-type annotation and inferred lineage trajectory onto this UMAP. (c) Single-cells whose expression profiles matched the CT gene signature (gene set UCell score > (median score + standard deviation)) are assigned to ‘6q-CT’ and shown in red; the remaining cells did not meet the threshold for the CT gene signature (assigned ‘6q-Ref’ status’). P-values depict enrichment of 6q-CT cells in clusters 3 and 7. (d) Significant enrichment of 6q-CT cells in clusters 3 and 7 based on scRNA-seq. Upper panel: A dot plot shows the significance of over-representation of 6q-CT calls in scRNA clusters based on targeted SCNA recalling (P-values based on FDR-adjusted Fisher’s exact tests). Lower panel: Gene set-level expression summary for the CT gene signature, which was derived using UCell81 with the directionality of expression changes taken into account. (e) Clonal frequency (CF) of 6q-CT cells after treatment with Notch inhibitor CB-103 (green) and vehicle control (brown) along a time course – 8h and 24h after treatment. CF was estimated by transferring gene set based CT annotations obtained from the scRNA-seq of T-ALL_P1 before treatment, to the scRNA-seq of T-ALL_P1 after treatment. Changed CF (%) at 24h compared to 8h is shown in the plot on top of the 24h data points. For each time point, the difference of CF under vehicle and CB-103 was evaluated by Fisher’s exact test (results are based on pairwise comparisons). (f) Scaled enrichment scores obtained by single-cell gene set enrichment analysis for the ‘N1-ICD transcriptional pathway’ gene set. Scores across treatment conditions (vehicle versus CB-103) were compared using two-sided Wilcoxon ranksum tests. (Boxplot was defined by minima=25th percentile-1.5X interquartile range (IQR), maxima=75th percentile+1.5X IQR, center=median, and bounds of box=25th and 75th percentile; n=665, 978, 915, and 556 cells for 6q-Ref from 8h Vehicle, 8h CB-103, 24h Vehicle, and 24h CB-103 (n=91, 157, 213, and 88) cells for 6q-CT for each condition respectively.) (g) Network representation of GOBPs enriched by differentially expressed genes in 6q-CT compared to 6q-Ref cells under CB-103 treatment (24h), subtracting any genes not specific to the drug treatment.