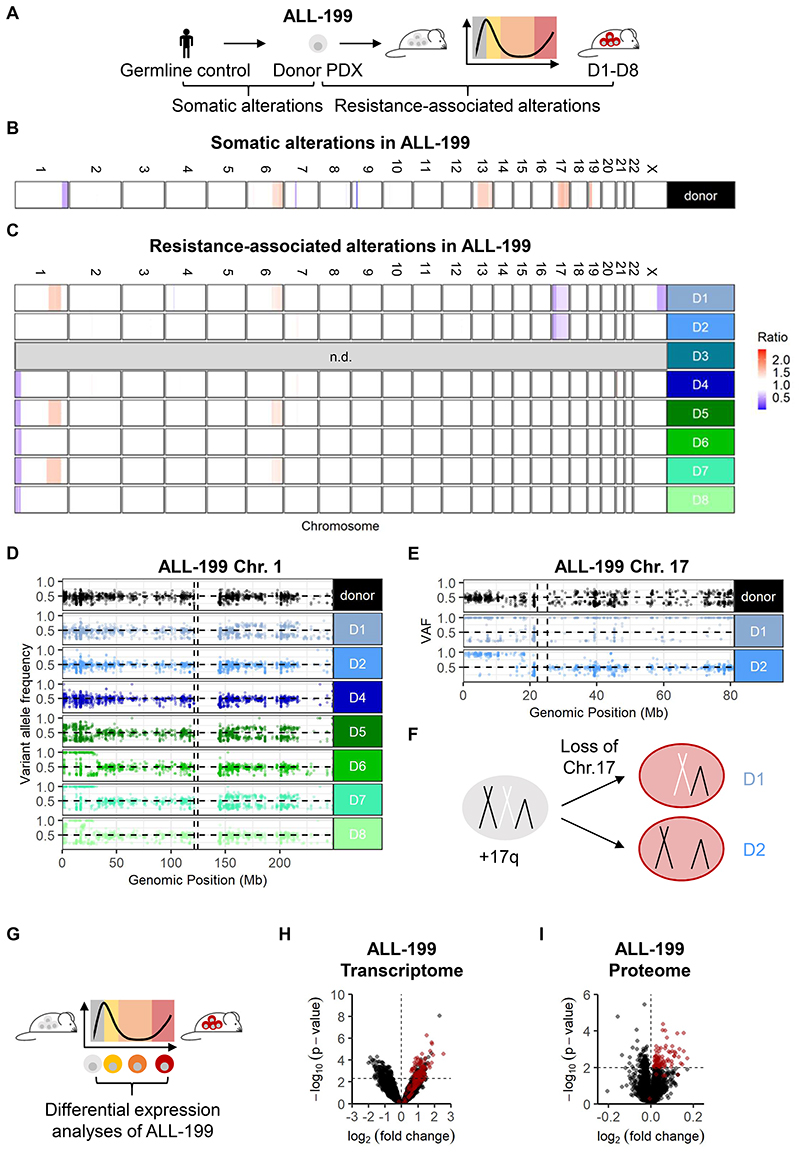
Figure 3. Heterogeneous genomic alterations, but shared expression features upon acquired resistance in ALL-199.
A-F Genomic characterization of resistant ALL-199 derivatives
A Whole exome sequencing was performed on ALL-199 germ line control obtained from healthy BM cells of the patient, donor PDX model, untreated cells and the eight resistant derivatives (D1-D8).
B-C Comparison between donor ALL-199 PDX and germline control to identify somatic alterations of the PDX leukemia (B); comparison between resistant D1-D8 and donor PDX to identify resistance-associated alterations (C); losses are depicted in blue and gains in red. Each row depicts one chromosome and each line depicts one sample. D3 was excluded due to poor sequencing quality.
D-E Magnification into chr. 1 (D) and chr. 17 (E); each dot represents a SNV as determined using hg19 as reference; homozygous SNV (VAF >0.8) present in the donor sample and SNV with VAF < 0.2 were excluded. Horizontal dashed line represents VAF = 0.5, vertical dashed lines represent centromere positions.
F Schematic representation of chr. 17 in donor PDX ALL-199 (light grey), where one allele of chr. 17q is duplicated, and in resistant derivatives D1 and D2 (red), where one of two whole chr. 17 alleles were lost.
G-I Transcriptomic and proteomic profiling
G Experimental layout: Transcriptomes were determined from ALL-199U (n=5), ALL-199S (n=6), ALL-199P (n=5) and ALL-199R (n=24, combined samples shown in Figures 1C, 2D and S2I). Proteomes of ALL-199U (n=4) and ALL-199R (n=8) were analyzed.
H Volcano plot representing genes differentially expressed between ALL-199U and ALL-199R; each dot represents one transcript; dashed lines indicate cut-offs for significant up- or downregulation (p < 0.005 and log2 fold-change > 0 or < 0, respectively). Red dots indicate candidates, which were chosen for further analysis.
I Volcano plot of all differentially expressed proteins between ALL-199U and ALL-199R is shown and depicted as in Figure 3H; dashed lines indicate cut-offs for significant up- or downregulation (p < 0.01 and log2 fold-change > 0 or < 0, respectively).