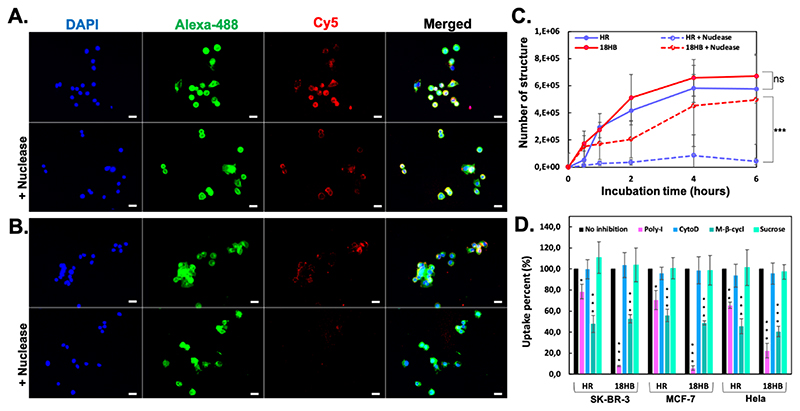
Figure 3. Cell uptake of DNA origami.
(A), with/without nuclease, fluorescence microscopy images showing Cy5-18HB (red) associated to SK-BR-3 cells. (B), with/without nuclease digestion, fluorescence microscopic images showing Cy5-HR (red) associated to SK-BR-3 cells. Cells were fixed and stained for nucleus using DAPI (blue) and membrane using WGA-Alexa-488 (green). (C), quantification of DNA origami by qPCR after incubating structures with SK-BR-3 cells. (D), qPCR analysis of the 4-hour uptake of DNA origami into SK-BR-3, MCF-7 and Hela cells with inhibition of scavenger receptors (Poly-I pretreatment), the non-receptor mediated endocytosis (CytoD pretreatment), the caveolin-dependent endocytosis (M-β-cycl pretreatment) or the clathrin-dependent endocytosis (Sucrose pretreatment). Each column represents three independent experiments. Data represents mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.