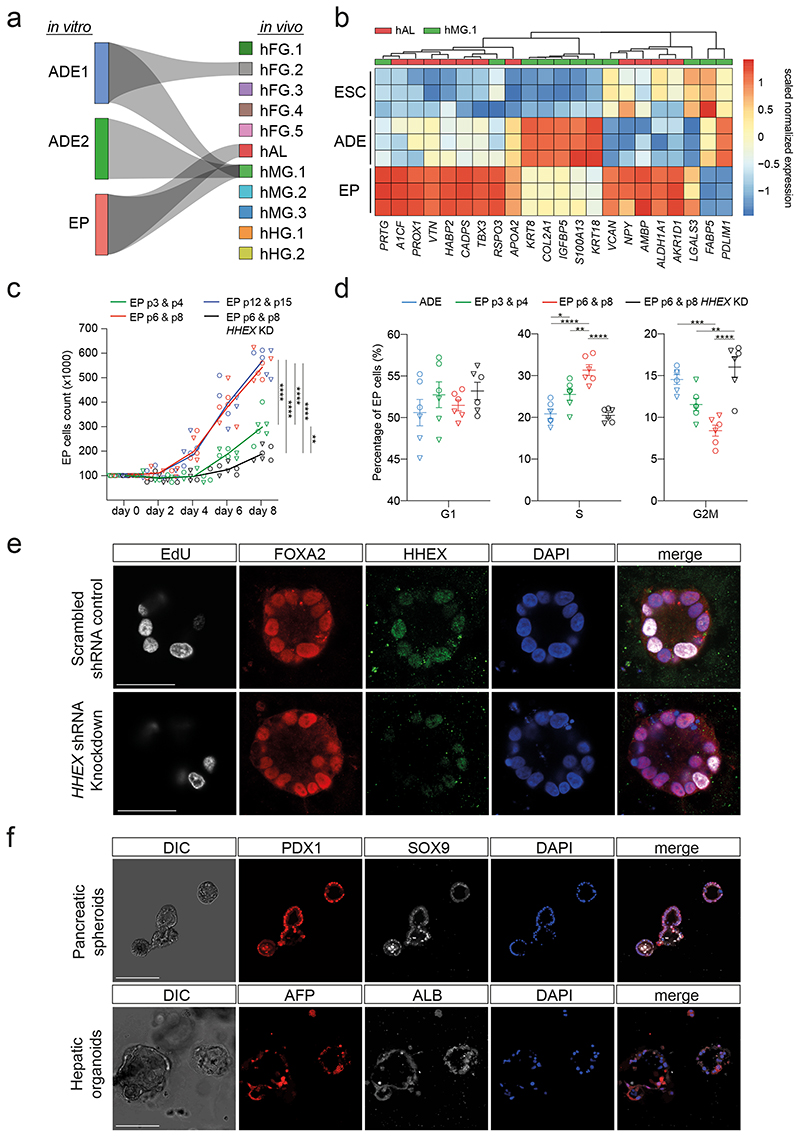
Fig. 1. Expanding endoderm progenitors as an in vitro model for ventral foregut.
a, Visualization of the CAT alignments between in vitro clusters (ADE.1, ADE.2, and EP) from this study and in vivo endodermal clusters from the Li et al. dataset20. Only significant CAT alignments between clusters are shown. b, Heatmap showing expression of hAL and hMG marker genes in ESC, ADE and EP cells (Bulk RNA-seq dataset, scaled normalized expression, N=3 independent experiments). Only markers expressed significantly different between ADE and EP are shown (log2 fold change > 1.5, adjusted P < 0.05). c, Cumulative growth curves showing EP cell counts at different passages of expansion for control and HHEX knockdown (EPs were derived from H9, circle, or HUES4, triangle, ESCs). Data are represented as mean ± SEM (N=6 independent experiments). **P <0.01, ****P <0.0001 (one-way ANOVA Tukey’s multiple comparison test was applied to analyze differences at day 8, only significant comparisons are shown). d, Dot plots showing percentage of G1, S, and G2M cycling cells assayed by flow cytometry with EdU and DAPI staining in control and HHEX knockdown EP expansion. Data are represented as mean ± SEM (N=6 independent experiments). *P <0.05, **P <0.01, ***P <0.001, ****P <0.0001 (oneway ANOVA Tukey’s multiple comparison test, only significant comparisons are shown). e, Representative images (from three independent experiments) of control (top row) and HHEX shRNA (bottom row) EP cells stained with EdU, FOXA2, HHEX, DAPI. Scale bar = 50μm. f, Top row: representative immunostaining of PDX1 and SOX9, including DAPI, of VFG-derived pancreatic spheroids at passage 5. Bottom row: representative immunostaining of AFP and ALB, including DAPI, of VFG-derived hepatic organoids at passage 5. Images represent three independent experiments. Scale bar = 50μm.