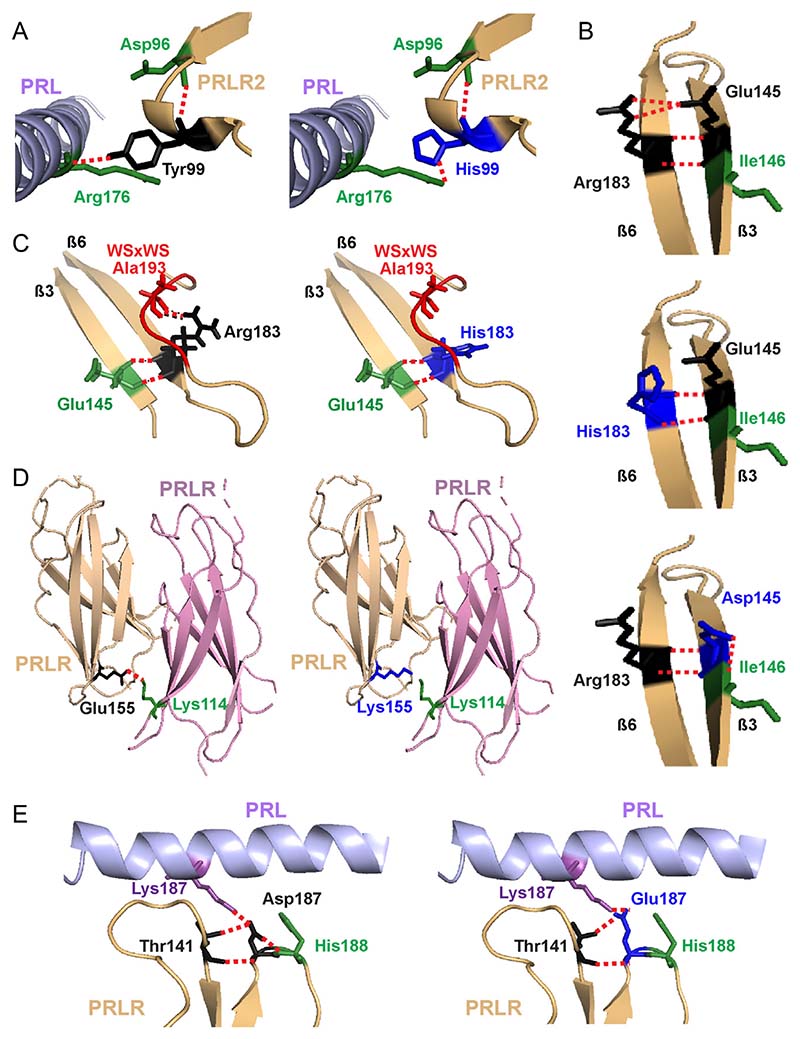
Figure 2. Structural characterisation of rare variants located in the C-terminal D1 and D2 of PRLR ECD.
(A) The Tyr99 residue is located close to the PRL-binding site with the PRLR. The hydroxyl group of the wild-type Tyr99 contacts Arg176 on the PRL protein (left). Mutation to His99 (blue, right) predicts retention of the contact, but at a more distal site, further away from the PRL α-helix and therefore is likely to increase the distance between the PRL and PRLR molecules which may affect binding and activation. (B) The Glu145 and Arg183 PRLR residues are located in adjacent β-strands and form polar contacts with each other. Mutation to His183 (middle) and Asp145 (bottom) is predicted to disrupt two of these contacts. Additionally, Asp145 is predicted to lose contact with the neighbouring Ile146, a residue previously demonstrated to be important for PRLR folding and stability (Dagil et al. 2012, Zhang et al. 2015). (C) The His183 also loses contact with Ala193, which forms part of the highly conserved WSxWS motif (red). (D) The wild-type Glu155 forms a contact with Lys114 on the opposite PRLR protomer. The Lys155 variant loses this contact and may disrupt homodimeric structural stability. (E) The wild-type Asp187 residue lies close to the PRL binding site and forms a contact with the His188 residue of PRLR, which has a critical role in ligand binding (Kulkarni et al. 2010) (Left). Mutation to Glu187 (blue, right) leads to loss of this contact, which may affect activation of the PRLR protein.