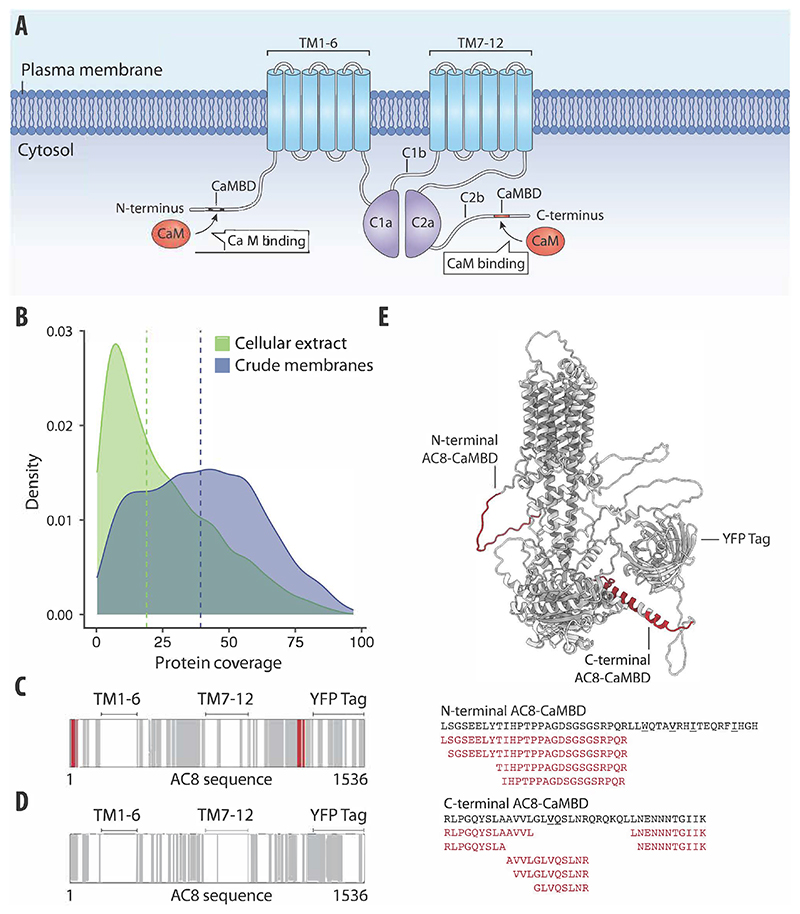
Figure 3. LiP–MS detects interactors of integral membrane proteins in crude membranes.
(A) Schematic of AC8 with the CaMBD in the N-terminus, transmembrane domains 1–6 and 7–12 (TM1–6 and TM7–12), and catalytic domains C1a, C1b, C2a, and C2b indicated. (B) Distribution of protein coverage for membrane-annotated proteins identified in crude membrane preparations of HEK293S GnTI-cells (blue) and in HEK293T cellular extracts (green). Blue and green vertical lines indicate calculated median coverages of 29.6% and 17.6%, respectively. (C, D) Protein sequence coverage of bovine AC8-YFP in LiP–MS in crude membranes (C) and in cellular extracts (D) is visualized. The barcodes depict peptides along the AC8-YFP sequence. Gray represents detected peptides, white represents non-detected regions, and red represents peptides that were significantly altered upon CaM addition (r > 0.85, |log2 FC|>1, moderated t-test, q value <0.01). (E) AlphaFold2 (Varadi et al, 2022; Jumper et al, 2021) predicted the structure of AC8 (including the tag domain) with peptides altered upon CaM addition, highlighted in red. Amino acid sequences comprising the CaM-binding motifs of AC8 are depicted in black. Hydrophobic residues of the CaM-binding motif are underlined. The significantly altered peptides upon CaM addition are shown in red.