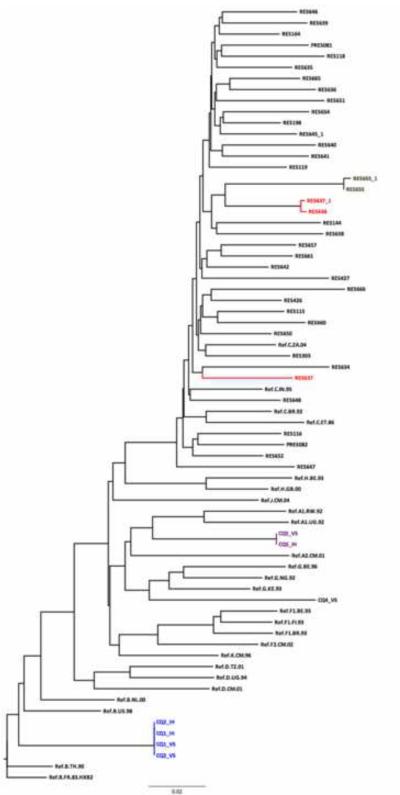
Figure 5. Use of a HKY Neighbor Joining tree done as part of sequence quality assurance.
There are four pairs/clusters of sequence with very short genetic distances. The genetic distance between RES655 and RES655_1 (same samples sequenced on different days) is 0.003. The is a potential error with the RES637_1/RES638 pair as their genetic distance is too short (0.075 ) for samples from different epidemiologically unlinked individuals. There is another RES637 on the tree with a distance of 0.075 when compared to RES638_1. The CQ01/CQ02 cluster suggests that the two samples from the panel are duplicates of the same sample. They cluster together with the subtype B reference sequence confirming the subtype assigned by the REGA Subtyping tool. CQ05 and CQ04 clustered with subtypes A and G respectively, whereas the REGA subtyping tool classified them as A and CRF02_AG respectively. Another useful tool for HIV subtyping and recombination is SCUEL, which is available at http://www.datamonkey.org.